| patient_id | trt | system_organ_class | adverse_event | grade | drug_attribution |
|---|---|---|---|---|---|
Introducing {gtreg}: an R package to produce regulatory tables for clinical research
Introduction
Developers
Checklist
Install recent R release
I am on version 4.2.1Install RStudio
I am on version 2022.12.0+353 Install packages
Ensure you can knit Rmarkdown files
Overview
gtreg

{gtreg} overview
About adverse events
Adverse events
An adverse event (AE) is a medical issue that occurs during the course of treatment or observation.
AEs are classified according to a hierarchy, and we typically report lower level terms within a system organ class.
AEs also record severity / grade and likelihood of attribution to treatment.
Summary table challenges
| patient_id | trt | system_organ_class | adverse_event | grade | drug_attribution |
|---|---|---|---|---|---|
| ID 1 | Drug B | Blood and lymphatic system disorders | Anaemia | 4 | Unrelated |
| ID 1 | Drug B | Blood and lymphatic system disorders | Increased tendency to bruise | 5 | Unrelated |
| ID 1 | Drug B | Blood and lymphatic system disorders | Increased tendency to bruise | 4 | Unlikely |
| ID 1 | Drug B | Blood and lymphatic system disorders | Thrombocytopenia | 1 | Probably |
| ID 1 | Drug B | Blood and lymphatic system disorders | Thrombocytopenia | 3 | Definite |
Subjects experience multiple adverse events.
Not all enrolled subjects experience an adverse event.
The percent of subjects experiencing specific AEs is of interest; typically AEs are counted by maximum grade per event per subject.
Multiple AE tables are often required (treatment emergent AEs, AEs on specific treatment cycles, serious AEs, etc.).
Counting by maximum grade
| patient_id | system_organ_class | adverse_event | grade |
|---|---|---|---|
| ID 1 | Blood and lymphatic system disorders | Anaemia | 4 |
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 5 |
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 4 |
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 1 |
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 3 |
Counting by maximum grade
| Patient ID | System Organ Class | Adverse Event | Grade |
|---|---|---|---|
| Term 1 | |||
| ID 1 | Blood and lymphatic system disorders | Anaemia | 4 |
| Term 2 | |||
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 5 |
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 4 |
| Term 3 | |||
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 1 |
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 3 |
Counting by maximum grade
| Patient ID | System Organ Class | Adverse Event | Grade |
|---|---|---|---|
| Term 1 | |||
| ID 1 | Blood and lymphatic system disorders | Anaemia | 4 |
| Term 2 | |||
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 5 |
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 4 |
| Term 3 | |||
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 1 |
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 3 |
Similar logic applies when counting by severity (e.g. mild, moderate, severe) - if stored as a factor, the highest factor level is retained (severe).
First adverse event tables
Data
df_adverse_events
10 unique subjects
multiple rows per subject
| patient_id | trt | system_organ_class | adverse_event | grade | drug_attribution | any_complication | grade3_complication |
|---|---|---|---|---|---|---|---|
| ID 1 | Drug B | Blood and lymphatic system disorders | Anaemia | 4 | Unrelated | TRUE | TRUE |
| ID 1 | Drug B | Blood and lymphatic system disorders | Increased tendency to bruise | 5 | Unrelated | TRUE | TRUE |
| ID 1 | Drug B | Blood and lymphatic system disorders | Increased tendency to bruise | 4 | Unlikely | TRUE | TRUE |
| ID 1 | Drug B | Blood and lymphatic system disorders | Thrombocytopenia | 1 | Probably | TRUE | FALSE |
| ID 1 | Drug B | Blood and lymphatic system disorders | Thrombocytopenia | 3 | Definite | TRUE | TRUE |
| ID 1 | Drug B | Gastrointestinal disorders | Intestinal dilatation | 2 | Definite | TRUE | FALSE |
df_patient_characteristics
100 unique subjects
1 row per subject
| patient_id | trt | age | marker | status | discontinued | off_trt_ae |
|---|---|---|---|---|---|---|
| ID 1 | Drug B | 28 | 4.807148 | Adverse Event | Yes | Intestinal dilatation |
| ID 2 | Drug B | 47 | 7.714544 | Completed Study | No | NA |
| ID 3 | Drug B | 39 | 6.449793 | Completed Study | No | NA |
| ID 4 | Drug B | 45 | 6.319944 | Subject Withdrew | Yes | NA |
| ID 5 | Drug B | 58 | 5.785283 | Physician Decision | Yes | NA |
| ID 6 | Drug A | 42 | 5.630501 | Completed Study | No | NA |
Variable labels
A variable label is an attribute of a variable in a data frame.
Where applicable, variable labels (not variable names) are printed in {gtsummary} and {gtreg} tables.
The example data in {gtreg} comes with variable labels; you can also create your own via
labelled::set_variable_labels().
First adverse event tables
tbl_ae_count()
- Count all AEs (not by maximum grade).
| Adverse Event | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Blood and lymphatic system disorders | 8 | 6 | 11 | 12 | 13 |
| Anaemia | 1 | — | 3 | 3 | 3 |
| Increased tendency to bruise | — | — | 2 | 5 | 2 |
| Iron deficiency anaemia | 4 | 3 | — | 3 | 3 |
| Thrombocytopenia | 3 | 3 | 6 | 1 | 5 |
| Gastrointestinal disorders | 14 | 9 | 7 | 11 | 9 |
| Difficult digestion | 1 | — | — | 3 | 1 |
| Intestinal dilatation | 3 | 1 | — | — | 1 |
| Myochosis | 1 | 4 | 3 | 2 | 3 |
| Non-erosive reflux disease | 5 | — | — | 4 | 3 |
| Pancreatic enzyme abnormality | 4 | 4 | 4 | 2 | 1 |
| adverse_event | system_organ_class | grade |
|---|---|---|
| Anaemia | Blood and lymphatic system disorders | 4 |
| Increased tendency to bruise | Blood and lymphatic system disorders | 5 |
| Increased tendency to bruise | Blood and lymphatic system disorders | 4 |
| Thrombocytopenia | Blood and lymphatic system disorders | 1 |
| Thrombocytopenia | Blood and lymphatic system disorders | 3 |
| Intestinal dilatation | Gastrointestinal disorders | 2 |
| Intestinal dilatation | Gastrointestinal disorders | 1 |
| Myochosis | Gastrointestinal disorders | 3 |
| Myochosis | Gastrointestinal disorders | 5 |
| Non-erosive reflux disease | Gastrointestinal disorders | 5 |
tbl_ae()
- Count AEs per subject by maximum grade.
| Adverse Event | N = 10 | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Blood and lymphatic system disorders | — | 1 (10) | — | 2 (20) | 7 (70) |
| Anaemia | — | — | 2 (20) | 2 (20) | 3 (30) |
| Increased tendency to bruise | — | — | — | 4 (40) | 2 (20) |
| Iron deficiency anaemia | 1 (10) | 2 (20) | — | 2 (20) | 2 (20) |
| Thrombocytopenia | — | 1 (10) | 3 (30) | 1 (10) | 4 (40) |
| Gastrointestinal disorders | — | — | — | 4 (40) | 6 (60) |
| Difficult digestion | 1 (10) | — | — | 3 (30) | 1 (10) |
| Intestinal dilatation | 2 (20) | 1 (10) | — | — | 1 (10) |
| Myochosis | — | 3 (30) | 1 (10) | 1 (10) | 3 (30) |
| Non-erosive reflux disease | 4 (40) | — | — | 3 (30) | 3 (30) |
| Pancreatic enzyme abnormality | 2 (20) | 1 (10) | 2 (20) | 2 (20) | 1 (10) |
| patient_id | adverse_event | system_organ_class | grade |
|---|---|---|---|
| ID 1 | Anaemia | Blood and lymphatic system disorders | 4 |
| ID 1 | Increased tendency to bruise | Blood and lymphatic system disorders | 5 |
| ID 1 | Increased tendency to bruise | Blood and lymphatic system disorders | 4 |
| ID 1 | Thrombocytopenia | Blood and lymphatic system disorders | 1 |
| ID 1 | Thrombocytopenia | Blood and lymphatic system disorders | 3 |
| ID 1 | Intestinal dilatation | Gastrointestinal disorders | 2 |
| ID 1 | Intestinal dilatation | Gastrointestinal disorders | 1 |
| ID 1 | Myochosis | Gastrointestinal disorders | 3 |
| ID 1 | Myochosis | Gastrointestinal disorders | 5 |
| ID 1 | Non-erosive reflux disease | Gastrointestinal disorders | 5 |
tbl_ae()
Count AEs per subject by maximum grade.
Supply
id_dfto achieve subject denominator.
| Adverse Event | N = 100 | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Blood and lymphatic system disorders | — | 1 (1.0) | — | 2 (2.0) | 7 (7.0) |
| Anaemia | — | — | 2 (2.0) | 2 (2.0) | 3 (3.0) |
| Increased tendency to bruise | — | — | — | 4 (4.0) | 2 (2.0) |
| Iron deficiency anaemia | 1 (1.0) | 2 (2.0) | — | 2 (2.0) | 2 (2.0) |
| Thrombocytopenia | — | 1 (1.0) | 3 (3.0) | 1 (1.0) | 4 (4.0) |
| Gastrointestinal disorders | — | — | — | 4 (4.0) | 6 (6.0) |
| Difficult digestion | 1 (1.0) | — | — | 3 (3.0) | 1 (1.0) |
| Intestinal dilatation | 2 (2.0) | 1 (1.0) | — | — | 1 (1.0) |
| Myochosis | — | 3 (3.0) | 1 (1.0) | 1 (1.0) | 3 (3.0) |
| Non-erosive reflux disease | 4 (4.0) | — | — | 3 (3.0) | 3 (3.0) |
| Pancreatic enzyme abnormality | 2 (2.0) | 1 (1.0) | 2 (2.0) | 2 (2.0) | 1 (1.0) |
df_adverse_events
| patient_id | adverse_event | system_organ_class | grade |
|---|---|---|---|
| ID 1 | Anaemia | Blood and lymphatic system disorders | 4 |
| ID 1 | Increased tendency to bruise | Blood and lymphatic system disorders | 5 |
| ID 1 | Increased tendency to bruise | Blood and lymphatic system disorders | 4 |
| ID 1 | Thrombocytopenia | Blood and lymphatic system disorders | 1 |
| ID 1 | Thrombocytopenia | Blood and lymphatic system disorders | 3 |
df_patient_characteristics
| patient_id | trt |
|---|---|
| ID 1 | Drug B |
| ID 2 | Drug B |
| ID 3 | Drug B |
| ID 4 | Drug B |
| ID 5 | Drug B |
tbl_ae() with strata
| Adverse Event | Drug A, N = 44 | Drug B, N = 56 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Blood and lymphatic system disorders | — | 1 (2.3) | — | 1 (2.3) | 1 (2.3) | — | — | — | 1 (1.8) | 6 (11) |
| Anaemia | — | — | 1 (2.3) | 1 (2.3) | — | — | — | 1 (1.8) | 1 (1.8) | 3 (5.4) |
| Increased tendency to bruise | — | — | — | 1 (2.3) | — | — | — | — | 3 (5.4) | 2 (3.6) |
| Iron deficiency anaemia | — | — | — | 1 (2.3) | 1 (2.3) | 1 (1.8) | 2 (3.6) | — | 1 (1.8) | 1 (1.8) |
| Thrombocytopenia | — | 1 (2.3) | — | 1 (2.3) | — | — | — | 3 (5.4) | — | 4 (7.1) |
| Gastrointestinal disorders | — | — | — | 2 (4.5) | 1 (2.3) | — | — | — | 2 (3.6) | 5 (8.9) |
| Difficult digestion | — | — | — | 3 (6.8) | — | 1 (1.8) | — | — | — | 1 (1.8) |
| Intestinal dilatation | 1 (2.3) | — | — | — | — | 1 (1.8) | 1 (1.8) | — | — | 1 (1.8) |
| Myochosis | — | 2 (4.5) | 1 (2.3) | — | — | — | 1 (1.8) | — | 1 (1.8) | 3 (5.4) |
| Non-erosive reflux disease | 3 (6.8) | — | — | — | — | 1 (1.8) | — | — | 3 (5.4) | 3 (5.4) |
| Pancreatic enzyme abnormality | — | — | 1 (2.3) | 1 (2.3) | 1 (2.3) | 2 (3.6) | 1 (1.8) | 1 (1.8) | 1 (1.8) | — |
df_adverse_events
| patient_id | trt | adverse_event | system_organ_class | grade |
|---|---|---|---|---|
| ID 1 | Drug B | Anaemia | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 5 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 1 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 3 |
df_patient_characteristics
| patient_id | trt |
|---|---|
| ID 1 | Drug B |
| ID 2 | Drug B |
| ID 3 | Drug B |
| ID 4 | Drug B |
| ID 5 | Drug B |
tbl_ae_focus()
Count dichotomous AE attributes via
include.Supply
id_dfto achieve subject denominator.
| Adverse Event | N = 100 | |
|---|---|---|
| Any Grade Complication | Grade 3+ Complication | |
| Blood and lymphatic system disorders | 10 (10) | 9 (9.0) |
| Anaemia | 7 (7.0) | 7 (7.0) |
| Increased tendency to bruise | 6 (6.0) | 6 (6.0) |
| Iron deficiency anaemia | 7 (7.0) | 4 (4.0) |
| Thrombocytopenia | 9 (9.0) | 8 (8.0) |
| Gastrointestinal disorders | 10 (10) | 10 (10) |
| Difficult digestion | 5 (5.0) | 4 (4.0) |
| Intestinal dilatation | 4 (4.0) | 1 (1.0) |
| Myochosis | 8 (8.0) | 5 (5.0) |
| Non-erosive reflux disease | 10 (10) | 6 (6.0) |
| Pancreatic enzyme abnormality | 8 (8.0) | 5 (5.0) |
df_adverse_events
| patient_id | adverse_event | system_organ_class | any_complication | grade3_complication |
|---|---|---|---|---|
| ID 1 | Anaemia | Blood and lymphatic system disorders | TRUE | TRUE |
| ID 1 | Increased tendency to bruise | Blood and lymphatic system disorders | TRUE | TRUE |
| ID 1 | Increased tendency to bruise | Blood and lymphatic system disorders | TRUE | TRUE |
| ID 1 | Thrombocytopenia | Blood and lymphatic system disorders | TRUE | FALSE |
| ID 1 | Thrombocytopenia | Blood and lymphatic system disorders | TRUE | TRUE |
df_patient_characteristics
| patient_id | trt |
|---|---|
| ID 1 | Drug B |
| ID 2 | Drug B |
| ID 3 | Drug B |
| ID 4 | Drug B |
| ID 5 | Drug B |
Modified adverse event tables
Modified adverse event tables
tbl_ae() with add_overall()
See documentation for more add_overall() functionality.
| Adverse Event | Drug A, N = 44 | Drug B, N = 56 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Overall | 1 | 2 | 3 | 4 | 5 | Overall | |
| Blood and lymphatic system disorders | — | 1 (2.3) | — | 1 (2.3) | 1 (2.3) | 3 (6.8) | — | — | — | 1 (1.8) | 6 (11) | 7 (12) |
| Anaemia | — | — | 1 (2.3) | 1 (2.3) | — | 2 (4.5) | — | — | 1 (1.8) | 1 (1.8) | 3 (5.4) | 5 (8.9) |
| Increased tendency to bruise | — | — | — | 1 (2.3) | — | 1 (2.3) | — | — | — | 3 (5.4) | 2 (3.6) | 5 (8.9) |
| Iron deficiency anaemia | — | — | — | 1 (2.3) | 1 (2.3) | 2 (4.5) | 1 (1.8) | 2 (3.6) | — | 1 (1.8) | 1 (1.8) | 5 (8.9) |
| Thrombocytopenia | — | 1 (2.3) | — | 1 (2.3) | — | 2 (4.5) | — | — | 3 (5.4) | — | 4 (7.1) | 7 (12) |
| Gastrointestinal disorders | — | — | — | 2 (4.5) | 1 (2.3) | 3 (6.8) | — | — | — | 2 (3.6) | 5 (8.9) | 7 (12) |
| Difficult digestion | — | — | — | 3 (6.8) | — | 3 (6.8) | 1 (1.8) | — | — | — | 1 (1.8) | 2 (3.6) |
| Intestinal dilatation | 1 (2.3) | — | — | — | — | 1 (2.3) | 1 (1.8) | 1 (1.8) | — | — | 1 (1.8) | 3 (5.4) |
| Myochosis | — | 2 (4.5) | 1 (2.3) | — | — | 3 (6.8) | — | 1 (1.8) | — | 1 (1.8) | 3 (5.4) | 5 (8.9) |
| Non-erosive reflux disease | 3 (6.8) | — | — | — | — | 3 (6.8) | 1 (1.8) | — | — | 3 (5.4) | 3 (5.4) | 7 (12) |
| Pancreatic enzyme abnormality | — | — | 1 (2.3) | 1 (2.3) | 1 (2.3) | 3 (6.8) | 2 (3.6) | 1 (1.8) | 1 (1.8) | 1 (1.8) | — | 5 (8.9) |
df_adverse_events
| patient_id | trt | adverse_event | system_organ_class | grade |
|---|---|---|---|---|
| ID 1 | Drug B | Anaemia | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 5 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 1 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 3 |
df_patient_characteristics
| patient_id | trt |
|---|---|
| ID 1 | Drug B |
| ID 2 | Drug B |
| ID 3 | Drug B |
| ID 4 | Drug B |
| ID 5 | Drug B |
tbl_ae() with modified headers
df_adverse_events |>
tbl_ae(
id = patient_id,
id_df = df_patient_characteristics,
strata = trt,
ae = adverse_event,
soc = system_organ_class,
by = grade
) |>
add_overall(across = 'by') |>
bold_labels() |>
modify_header(
all_ae_cols() ~ "**Grade {by}**"
) %>%
modify_spanning_header(
all_ae_cols(TRUE, TRUE) ~ "**{strata}** \nN = {n}"
)| Adverse Event |
Drug A N = 44 |
Drug B N = 56 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | Overall | Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | Overall | |
| Blood and lymphatic system disorders | — | 1 (2.3) | — | 1 (2.3) | 1 (2.3) | 3 (6.8) | — | — | — | 1 (1.8) | 6 (11) | 7 (12) |
| Anaemia | — | — | 1 (2.3) | 1 (2.3) | — | 2 (4.5) | — | — | 1 (1.8) | 1 (1.8) | 3 (5.4) | 5 (8.9) |
| Increased tendency to bruise | — | — | — | 1 (2.3) | — | 1 (2.3) | — | — | — | 3 (5.4) | 2 (3.6) | 5 (8.9) |
| Iron deficiency anaemia | — | — | — | 1 (2.3) | 1 (2.3) | 2 (4.5) | 1 (1.8) | 2 (3.6) | — | 1 (1.8) | 1 (1.8) | 5 (8.9) |
| Thrombocytopenia | — | 1 (2.3) | — | 1 (2.3) | — | 2 (4.5) | — | — | 3 (5.4) | — | 4 (7.1) | 7 (12) |
| Gastrointestinal disorders | — | — | — | 2 (4.5) | 1 (2.3) | 3 (6.8) | — | — | — | 2 (3.6) | 5 (8.9) | 7 (12) |
| Difficult digestion | — | — | — | 3 (6.8) | — | 3 (6.8) | 1 (1.8) | — | — | — | 1 (1.8) | 2 (3.6) |
| Intestinal dilatation | 1 (2.3) | — | — | — | — | 1 (2.3) | 1 (1.8) | 1 (1.8) | — | — | 1 (1.8) | 3 (5.4) |
| Myochosis | — | 2 (4.5) | 1 (2.3) | — | — | 3 (6.8) | — | 1 (1.8) | — | 1 (1.8) | 3 (5.4) | 5 (8.9) |
| Non-erosive reflux disease | 3 (6.8) | — | — | — | — | 3 (6.8) | 1 (1.8) | — | — | 3 (5.4) | 3 (5.4) | 7 (12) |
| Pancreatic enzyme abnormality | — | — | 1 (2.3) | 1 (2.3) | 1 (2.3) | 3 (6.8) | 2 (3.6) | 1 (1.8) | 1 (1.8) | 1 (1.8) | — | 5 (8.9) |
df_adverse_events
| patient_id | trt | adverse_event | system_organ_class | grade |
|---|---|---|---|---|
| ID 1 | Drug B | Anaemia | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 5 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 1 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 3 |
df_patient_characteristics
| patient_id | trt |
|---|---|
| ID 1 | Drug B |
| ID 2 | Drug B |
| ID 3 | Drug B |
| ID 4 | Drug B |
| ID 5 | Drug B |
Table modifications
https://shannonpileggi.github.io/gtreg/articles/table-modifications.html
Use modify_header() and modify_spanning_header() with all_ column selectors.
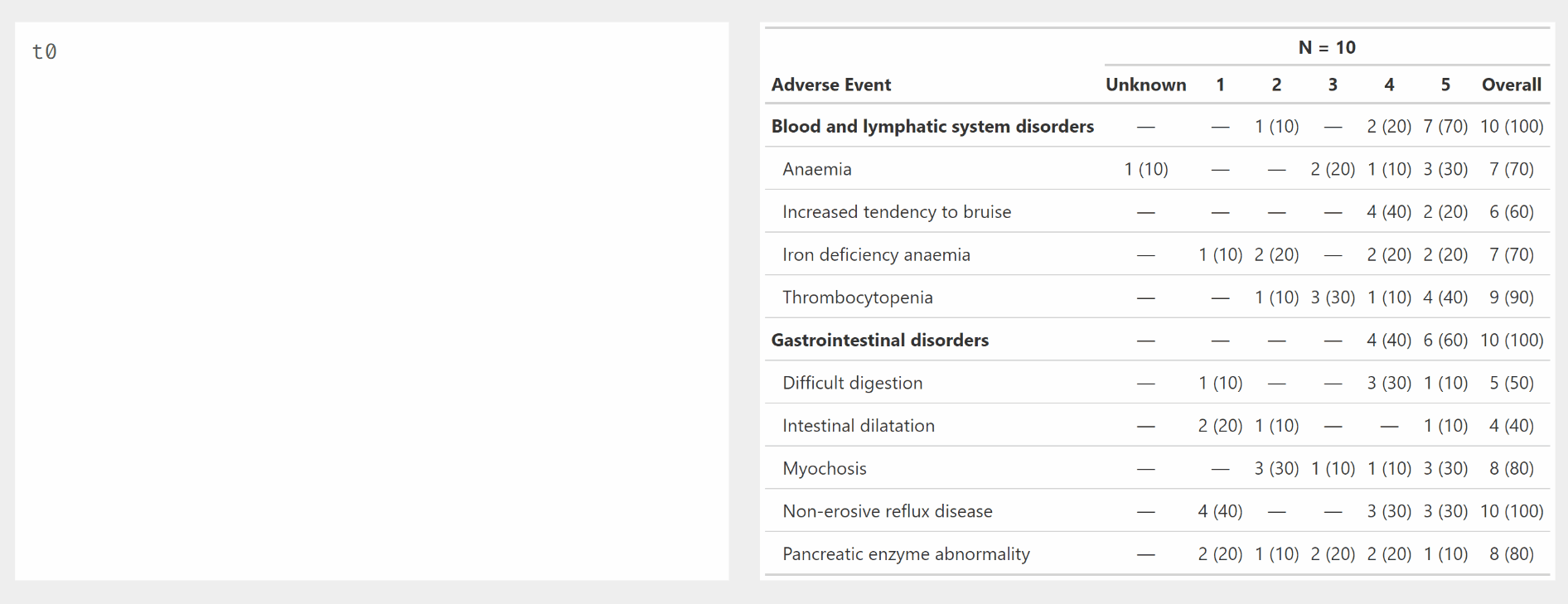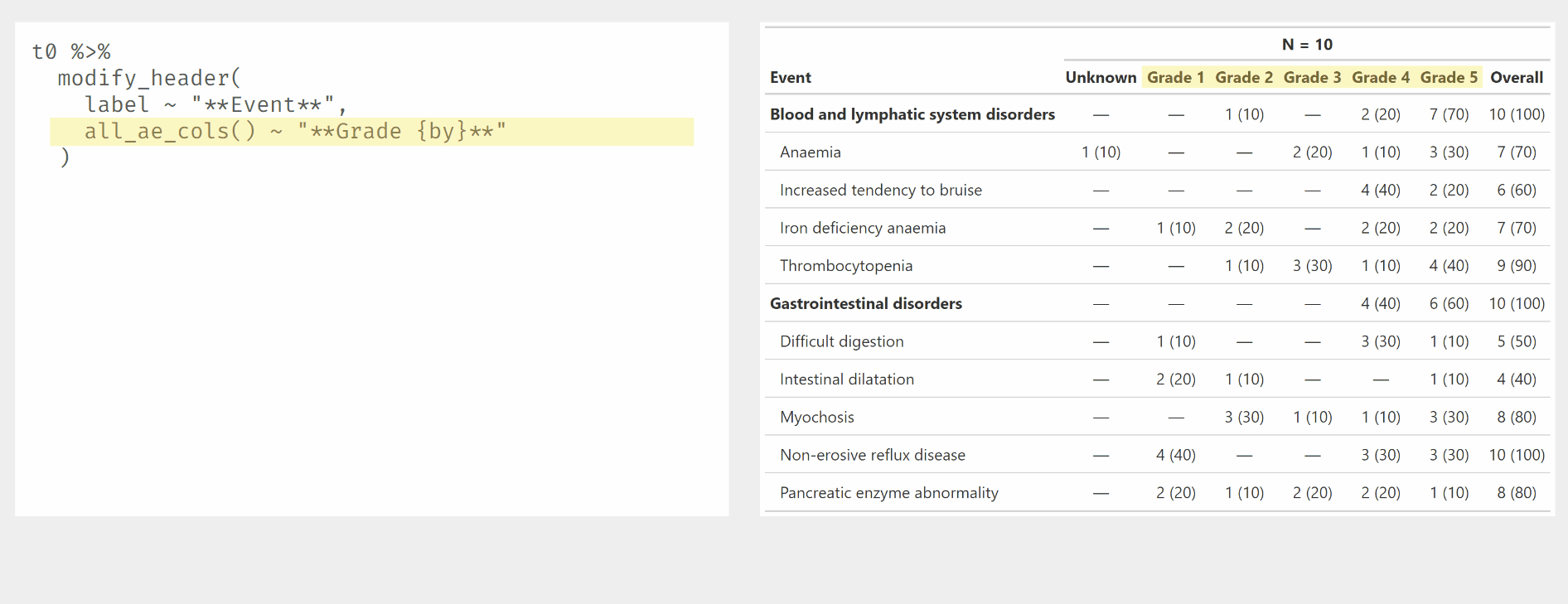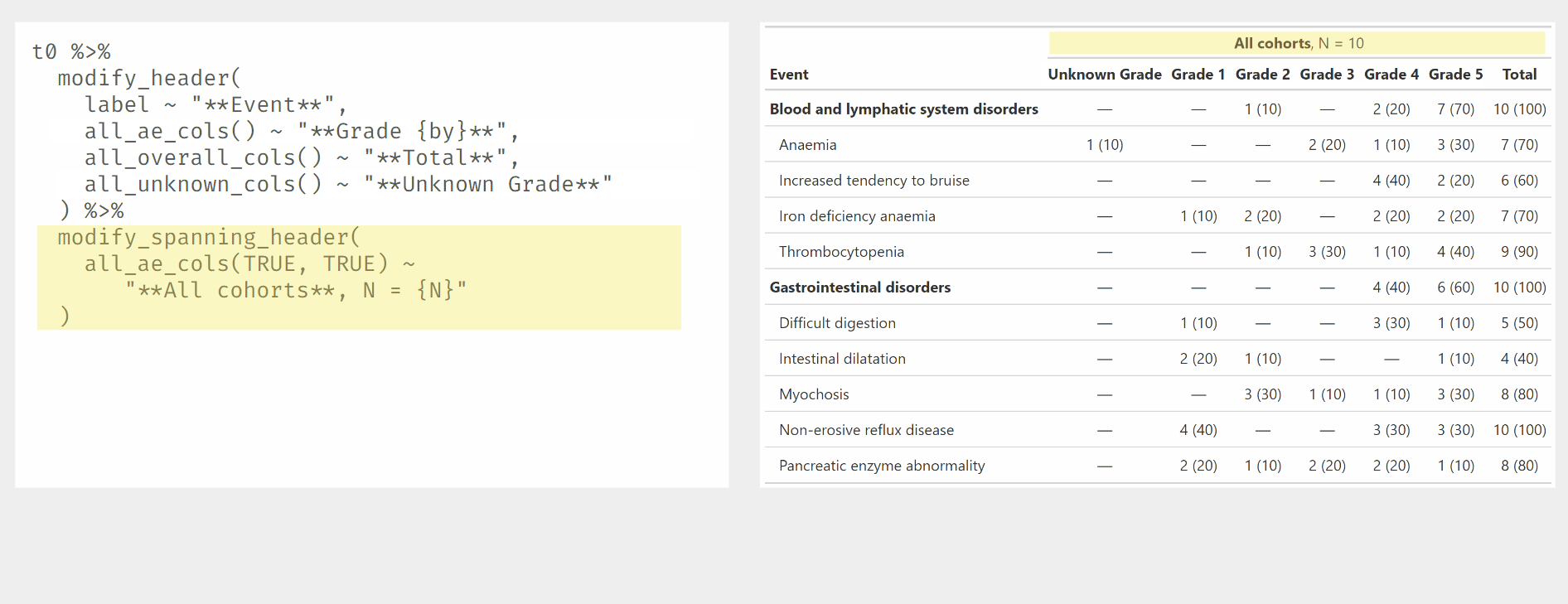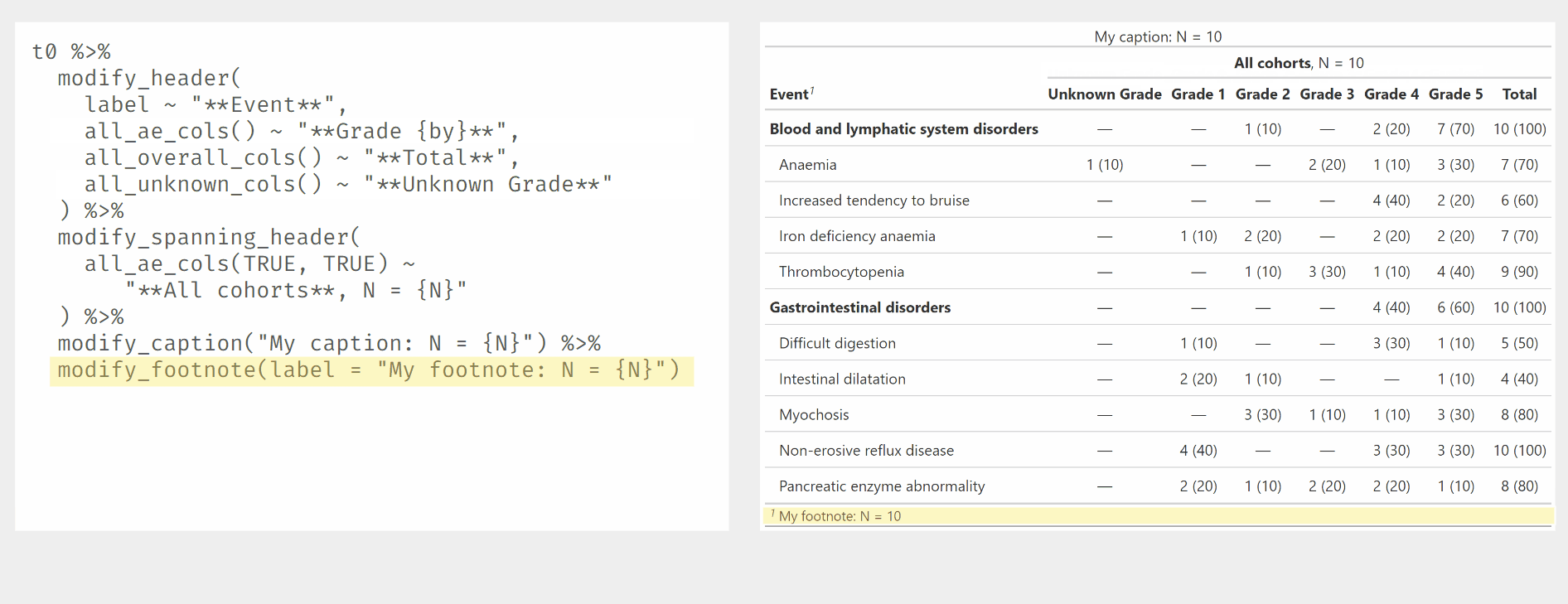
Other tabling functions
Other tabling functions
tbl_reg_summary()
Create summary tables with standard regulatory formatting.
| Characteristic | Drug A, N = 441 | Drug B, N = 561 |
|---|---|---|
| Biological Marker | ||
| N | 44 | 56 |
| Mean (SD) | 5.47 (0.97) | 5.46 (1.04) |
| Median (IQR) | 5.27 (4.98, 5.91) | 5.43 (4.67, 6.10) |
| Range | 3.53, 8.42 | 3.66, 8.00 |
| N missing | 0 | 0 |
| Study Status | ||
| Completed Study | 20 (45%) | 29 (52%) |
| Adverse Event | 4 (9.1%) | 10 (18%) |
| Progressive Disease | 4 (9.1%) | 4 (7.1%) |
| Physician Decision | 5 (11%) | 3 (5.4%) |
| Subject Withdrew | 4 (9.1%) | 6 (11%) |
| Active | 7 (16%) | 4 (7.1%) |
| 1 n (%) | ||
df_patient_characteristics
| patient_id | trt | marker | status |
|---|---|---|---|
| ID 1 | Drug B | 4.807148 | Adverse Event |
| ID 2 | Drug B | 7.714544 | Completed Study |
| ID 3 | Drug B | 6.449793 | Completed Study |
| ID 4 | Drug B | 6.319944 | Subject Withdrew |
| ID 5 | Drug B | 5.785283 | Physician Decision |
tbl_listing()
A fancy 💅 way to print grouped data.
| Adverse Event | Grade | Drug Attribution | Patient ID |
|---|---|---|---|
| Blood and lymphatic system disorders | |||
| Anaemia | 4 | Unrelated | ID 1 |
| Increased tendency to bruise | 5 | Unrelated | ID 1 |
| Increased tendency to bruise | 4 | Unlikely | ID 1 |
| Thrombocytopenia | 3 | Definite | ID 1 |
| Thrombocytopenia | 1 | Probably | ID 1 |
| Gastrointestinal disorders | |||
| Intestinal dilatation | 2 | Definite | ID 1 |
| Intestinal dilatation | 1 | Definite | ID 1 |
| Myochosis | 5 | Probably | ID 1 |
| Myochosis | 3 | Unlikely | ID 1 |
| Non-erosive reflux disease | 5 | Unrelated | ID 1 |
df_adverse_events
| system_organ_class | adverse_event | grade | drug_attribution | patient_id |
|---|---|---|---|---|
| Blood and lymphatic system disorders | Anaemia | 4 | Unrelated | ID 1 |
| Blood and lymphatic system disorders | Increased tendency to bruise | 5 | Unrelated | ID 1 |
| Blood and lymphatic system disorders | Increased tendency to bruise | 4 | Unlikely | ID 1 |
| Gastrointestinal disorders | Intestinal dilatation | 2 | Definite | ID 1 |
| Gastrointestinal disorders | Intestinal dilatation | 1 | Definite | ID 1 |
| Gastrointestinal disorders | Myochosis | 5 | Probably | ID 1 |
| Gastrointestinal disorders | Myochosis | 3 | Unlikely | ID 1 |
| Gastrointestinal disorders | Non-erosive reflux disease | 5 | Unrelated | ID 1 |
| Blood and lymphatic system disorders | Thrombocytopenia | 3 | Definite | ID 1 |
| Blood and lymphatic system disorders | Thrombocytopenia | 1 | Probably | ID 1 |
tbl_listing() extensions
Remember this table?
| Patient ID | System Organ Class | Adverse Event | Grade |
|---|---|---|---|
| Term 1 | |||
| ID 1 | Blood and lymphatic system disorders | Anaemia | 4 |
| Term 2 | |||
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 5 |
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 4 |
| Term 3 | |||
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 1 |
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 3 |
| patient_id | system_organ_class | adverse_event | grade | ae_label |
|---|---|---|---|---|
| ID 1 | Blood and lymphatic system disorders | Anaemia | 4 | Term 1 |
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 5 | Term 2 |
| ID 1 | Blood and lymphatic system disorders | Increased tendency to bruise | 4 | Term 2 |
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 1 | Term 3 |
| ID 1 | Blood and lymphatic system disorders | Thrombocytopenia | 3 | Term 3 |
Table shells
Table shells
Strategy
https://shannonpileggi.github.io/gtreg/articles/table-shells.html
Table shells can be generated for any tbl_ae() and tbl_reg_summary() by:
Create dummy data or use your own data.
Pass the data to your function of choice.
Overwrite the statistic(s) shown to a fixed character string by implementing the
style_xxx()function in thedigitsargument.
Uniform shell for tbl_ae()
df_adverse_events |>
tbl_ae(
id = patient_id,
id_df = df_patient_characteristics,
strata = trt,
ae = adverse_event,
soc = system_organ_class,
by = grade,
digits = style_xxx,
zero_symbol = NULL
) |>
bold_labels() |>
modify_header(all_ae_cols() ~ "**Grade {by}**") %>%
modify_spanning_header(all_ae_cols(TRUE, TRUE) ~ "**{strata}** \nN = xx")| Adverse Event |
Drug A N = xx |
Drug B N = xx |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | |
| Blood and lymphatic system disorders | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Anaemia | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Increased tendency to bruise | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Iron deficiency anaemia | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Thrombocytopenia | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Gastrointestinal disorders | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Difficult digestion | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Intestinal dilatation | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Myochosis | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Non-erosive reflux disease | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
| Pancreatic enzyme abnormality | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) | xx (xx) |
df_adverse_events
| patient_id | trt | adverse_event | system_organ_class | grade |
|---|---|---|---|---|
| ID 1 | Drug B | Anaemia | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 5 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 1 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 3 |
df_patient_characteristics
| patient_id | trt |
|---|---|
| ID 1 | Drug B |
| ID 2 | Drug B |
| ID 3 | Drug B |
| ID 4 | Drug B |
| ID 5 | Drug B |
Custom shell for tbl_ae()
df_adverse_events |>
tbl_ae(
id = patient_id,
id_df = df_patient_characteristics,
strata = trt,
ae = adverse_event,
soc = system_organ_class,
by = grade,
digits = list(
style_xxx, # style for n
function(x) style_xxx(x, width = 4, digits = 1) # style for %
),
zero_symbol = NULL
) |>
bold_labels() |>
modify_header(all_ae_cols() ~ "**Grade {by}**") %>%
modify_spanning_header(all_ae_cols(TRUE, TRUE) ~ "**{strata}** \nN = xx")| Adverse Event |
Drug A N = xx |
Drug B N = xx |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | |
| Blood and lymphatic system disorders | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Anaemia | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Increased tendency to bruise | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Iron deficiency anaemia | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Thrombocytopenia | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Gastrointestinal disorders | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Difficult digestion | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Intestinal dilatation | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Myochosis | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Non-erosive reflux disease | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
| Pancreatic enzyme abnormality | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) | xx (xx.x) |
df_adverse_events
| patient_id | trt | adverse_event | system_organ_class | grade |
|---|---|---|---|---|
| ID 1 | Drug B | Anaemia | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 5 |
| ID 1 | Drug B | Increased tendency to bruise | Blood and lymphatic system disorders | 4 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 1 |
| ID 1 | Drug B | Thrombocytopenia | Blood and lymphatic system disorders | 3 |
df_patient_characteristics
| patient_id | trt |
|---|---|
| ID 1 | Drug B |
| ID 2 | Drug B |
| ID 3 | Drug B |
| ID 4 | Drug B |
| ID 5 | Drug B |
Exporting options
Exporting options
https://shannonpileggi.github.io/gtreg/articles/output-gtreg.html
Exporting options
| Adverse Event | N = 100 | ||||
|---|---|---|---|---|---|
| Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | |
| Blood and lymphatic system disorders | — | 1 (1.0) | — | 2 (2.0) | 7 (7.0) |
| Anaemia | — | — | 2 (2.0) | 2 (2.0) | 3 (3.0) |
| Increased tendency to bruise | — | — | — | 4 (4.0) | 2 (2.0) |
| Iron deficiency anaemia | 1 (1.0) | 2 (2.0) | — | 2 (2.0) | 2 (2.0) |
| Thrombocytopenia | — | 1 (1.0) | 3 (3.0) | 1 (1.0) | 4 (4.0) |
| Gastrointestinal disorders | — | — | — | 4 (4.0) | 6 (6.0) |
| Difficult digestion | 1 (1.0) | — | — | 3 (3.0) | 1 (1.0) |
| Intestinal dilatation | 2 (2.0) | 1 (1.0) | — | — | 1 (1.0) |
| Myochosis | — | 3 (3.0) | 1 (1.0) | 1 (1.0) | 3 (3.0) |
| Non-erosive reflux disease | 4 (4.0) | — | — | 3 (3.0) | 3 (3.0) |
| Pancreatic enzyme abnormality | 2 (2.0) | 1 (1.0) | 2 (2.0) | 2 (2.0) | 1 (1.0) |
Thank you!
{gtreg} website
https://shannonpileggi.github.io/gtreg/
{gtreg} installation
{gtreg} issues
https://github.com/shannonpileggi/gtreg/issues
{gtreg} feature requests or questions: ask on https://github.com/shannonpileggi/gtreg/discussions


