
Table modifications: headers, footnotes, and captions
Source:vignettes/articles/table-modifications.Rmd
table-modifications.RmdOverview
In general, use modify_header() and
modify_spanning_header() to specify how headers
should be modified; use in conjunction with column selectors to specify
which column headers to modify. Use
show_header_names() on a saved table object to display a
usage guide for modifying headers.
To specify footnotes and captions, use modify_footnote()
and modify_caption().
For all modify_ functions, formatting is applied via
markdown and glue syntax can be used to insert summary statistics. In
addition, a return carriage inserts a line break when preceded by two
spaces: \n.
AE tables
For adverse event tables functions tbl_ae(),
tbl_ae_focus(), and tbl_ae_count(), use the
{gtreg} column selectors all_ae_cols(),
all_overall_cols(), all_unknown_cols(), and
all_cols_in_strata() to specify which columns to
apply formatting.
| {gtreg} column selectors and resulting columns selected. | |||
| Selector | AE | Overall | Unknown |
|---|---|---|---|
all_ae_cols() |
✔️ | ||
all_overall_cols() |
✔️ | ||
all_unknown_cols() |
✔️ | ||
all_ae_cols(overall = TRUE, unknown = TRUE) |
✔️ | ✔️ | ✔️ |
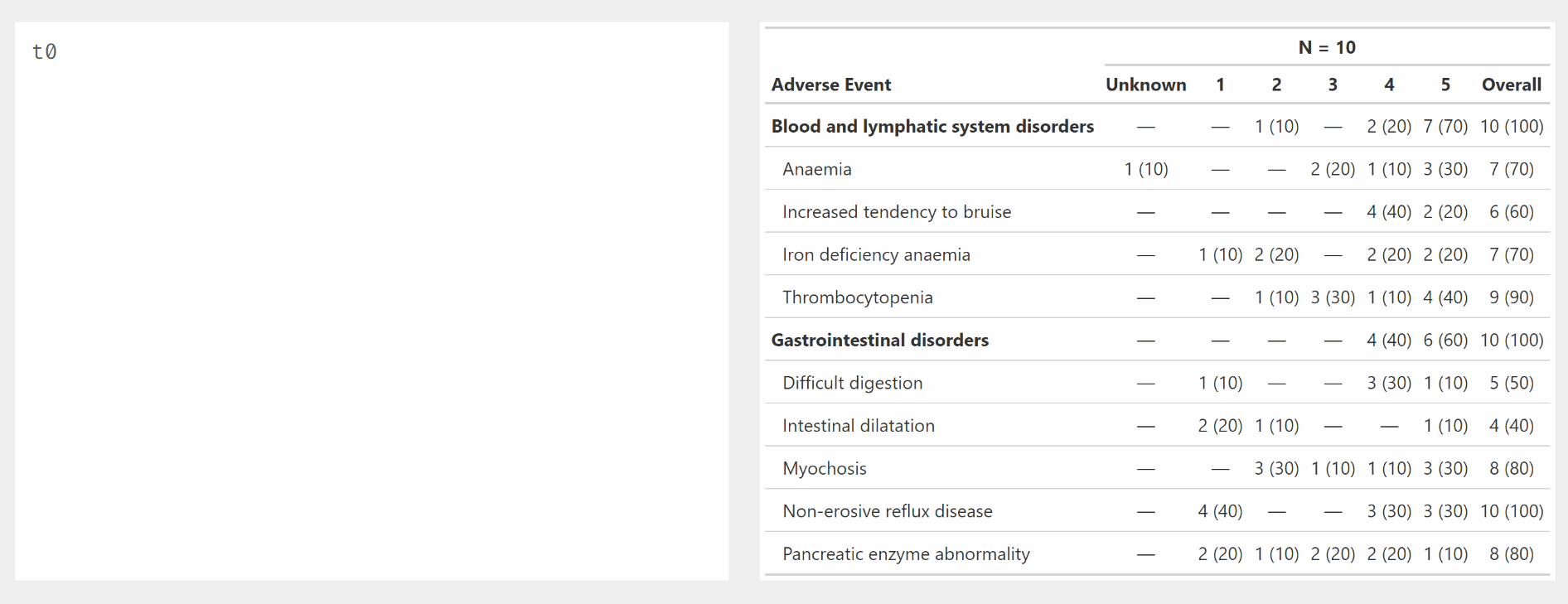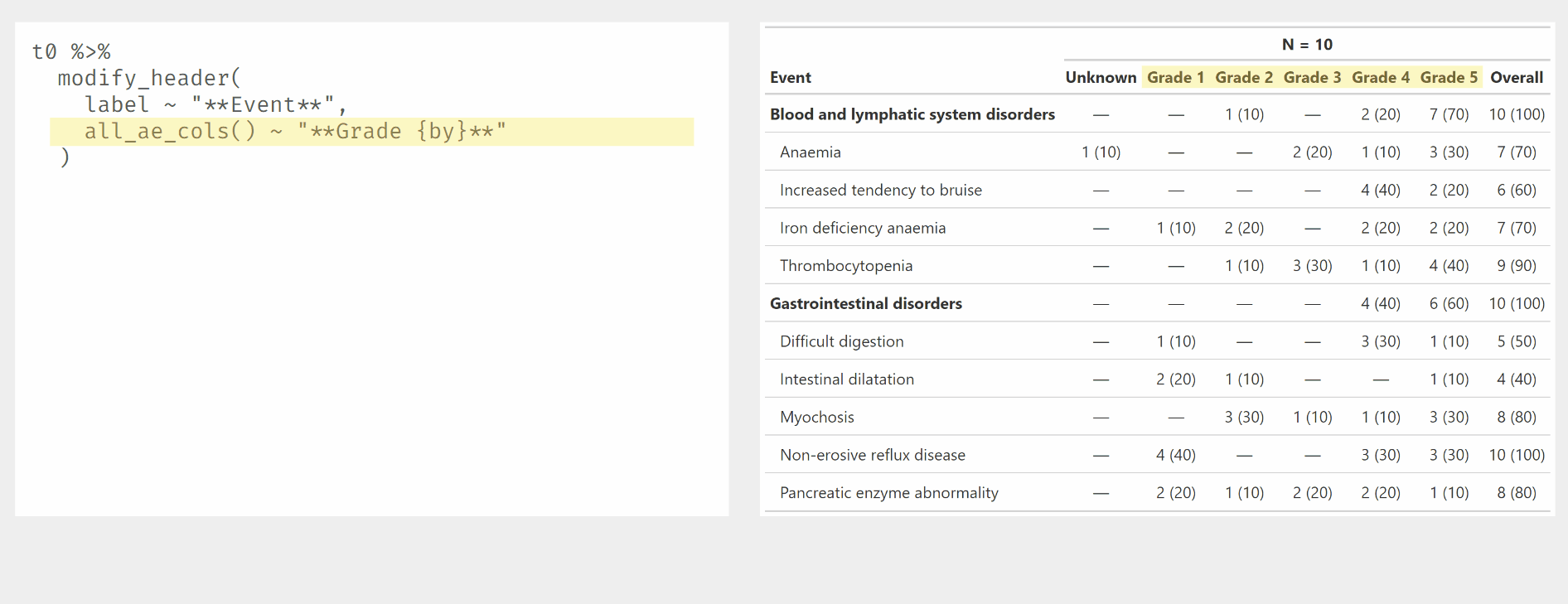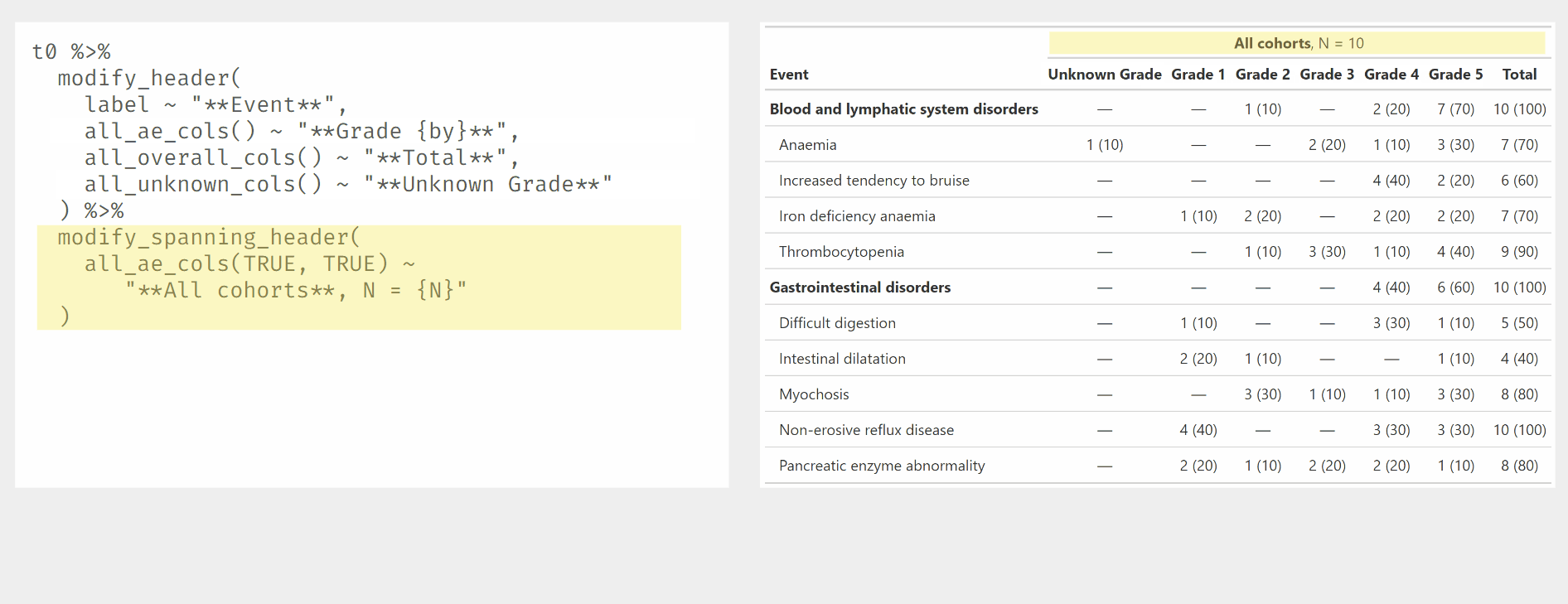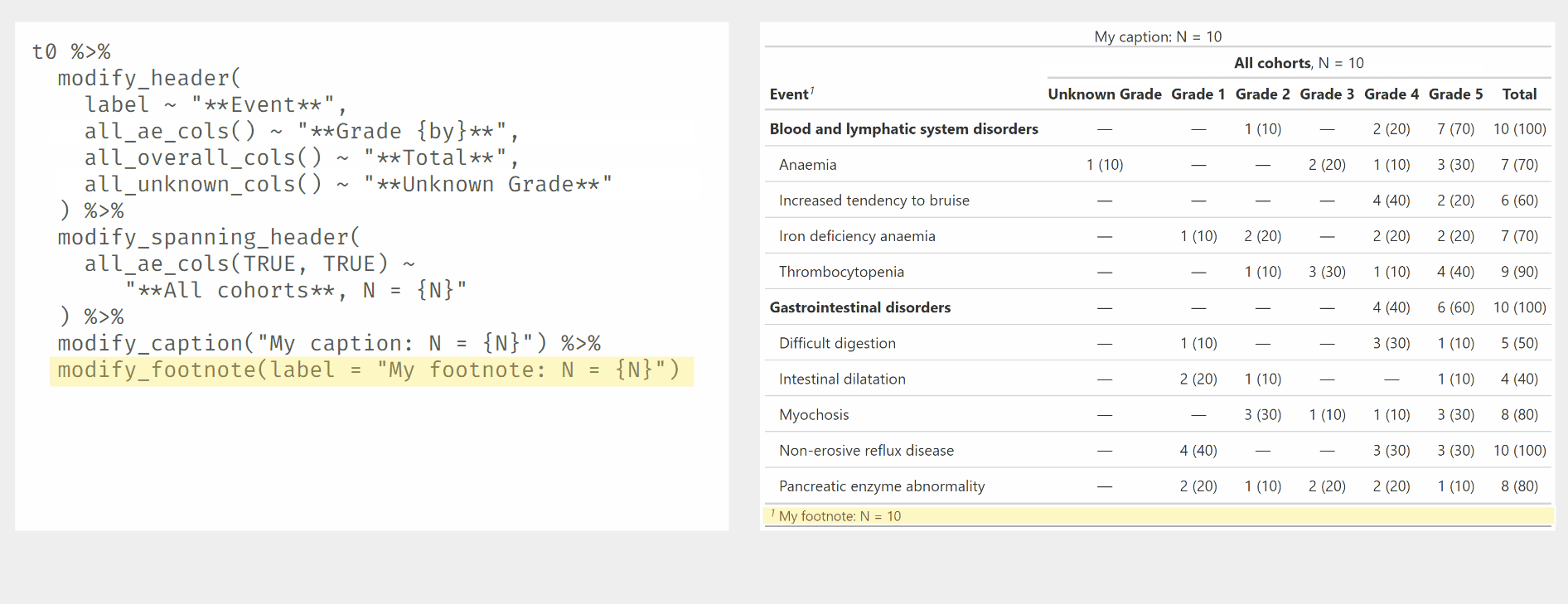
Demonstration of modify_ functions used with {gtreg} column
selectors.
Additionally, the all_cols_in_strata() selector can be
used with modify_spanning_header() to apply differing
modifications within strata.
tbl_ae() without strata
tbl1 <- df_adverse_events %>%
# create a missing value to demonstrate unknown columns
mutate(grade = ifelse(dplyr::row_number() == 1L, NA, grade)) %>%
tbl_ae(
id = patient_id,
ae = adverse_event,
soc = system_organ_class,
by = grade
) %>%
add_overall(across = 'by') %>%
bold_labels()Unmodified
# show_header_names(tbl1)
tbl1| Adverse Event |
N = 10
|
||||||
|---|---|---|---|---|---|---|---|
| Unknown | 1 | 2 | 3 | 4 | 5 | Overall | |
| Blood and lymphatic system disorders | — | — | 1 (10) | — | 2 (20) | 7 (70) | 10 (100) |
| Anaemia | 1 (10) | — | — | 2 (20) | 1 (10) | 3 (30) | 7 (70) |
| Increased tendency to bruise | — | — | — | — | 4 (40) | 2 (20) | 6 (60) |
| Iron deficiency anaemia | — | 1 (10) | 2 (20) | — | 2 (20) | 2 (20) | 7 (70) |
| Thrombocytopenia | — | — | 1 (10) | 3 (30) | 1 (10) | 4 (40) | 9 (90) |
| Gastrointestinal disorders | — | — | — | — | 4 (40) | 6 (60) | 10 (100) |
| Difficult digestion | — | 1 (10) | — | — | 3 (30) | 1 (10) | 5 (50) |
| Intestinal dilatation | — | 2 (20) | 1 (10) | — | — | 1 (10) | 4 (40) |
| Myochosis | — | — | 3 (30) | 1 (10) | 1 (10) | 3 (30) | 8 (80) |
| Non-erosive reflux disease | — | 4 (40) | — | — | 3 (30) | 3 (30) | 10 (100) |
| Pancreatic enzyme abnormality | — | 2 (20) | 1 (10) | 2 (20) | 2 (20) | 1 (10) | 8 (80) |
Modified
tbl1 %>%
modify_header(
label ~ "**Event**",
all_ae_cols() ~ "**Grade {by}**",
all_overall_cols() ~ "**Total**",
all_unknown_cols() ~ "**Unknown Grade**"
) %>%
modify_spanning_header(
all_ae_cols(TRUE, TRUE) ~ "**All cohorts**, N = {N}"
) |
All cohorts, N = 10
|
|||||||
|---|---|---|---|---|---|---|---|
| Event | Unknown Grade | Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | Total |
| Blood and lymphatic system disorders | — | — | 1 (10) | — | 2 (20) | 7 (70) | 10 (100) |
| Anaemia | 1 (10) | — | — | 2 (20) | 1 (10) | 3 (30) | 7 (70) |
| Increased tendency to bruise | — | — | — | — | 4 (40) | 2 (20) | 6 (60) |
| Iron deficiency anaemia | — | 1 (10) | 2 (20) | — | 2 (20) | 2 (20) | 7 (70) |
| Thrombocytopenia | — | — | 1 (10) | 3 (30) | 1 (10) | 4 (40) | 9 (90) |
| Gastrointestinal disorders | — | — | — | — | 4 (40) | 6 (60) | 10 (100) |
| Difficult digestion | — | 1 (10) | — | — | 3 (30) | 1 (10) | 5 (50) |
| Intestinal dilatation | — | 2 (20) | 1 (10) | — | — | 1 (10) | 4 (40) |
| Myochosis | — | — | 3 (30) | 1 (10) | 1 (10) | 3 (30) | 8 (80) |
| Non-erosive reflux disease | — | 4 (40) | — | — | 3 (30) | 3 (30) | 10 (100) |
| Pancreatic enzyme abnormality | — | 2 (20) | 1 (10) | 2 (20) | 2 (20) | 1 (10) | 8 (80) |
tbl_ae() with strata
tbl2 <-
df_adverse_events %>%
tbl_ae(
id = patient_id,
soc = system_organ_class,
ae = adverse_event,
strata = trt,
by = grade
) %>%
bold_labels()Unmodified
# show_header_names(tbl2)
tbl2| Adverse Event |
Drug A, N = 3
|
Drug B, N = 7
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Blood and lymphatic system disorders | — | 1 (33) | — | 1 (33) | 1 (33) | — | — | — | 1 (14) | 6 (86) |
| Anaemia | — | — | 1 (33) | 1 (33) | — | — | — | 1 (14) | 1 (14) | 3 (43) |
| Increased tendency to bruise | — | — | — | 1 (33) | — | — | — | — | 3 (43) | 2 (29) |
| Iron deficiency anaemia | — | — | — | 1 (33) | 1 (33) | 1 (14) | 2 (29) | — | 1 (14) | 1 (14) |
| Thrombocytopenia | — | 1 (33) | — | 1 (33) | — | — | — | 3 (43) | — | 4 (57) |
| Gastrointestinal disorders | — | — | — | 2 (67) | 1 (33) | — | — | — | 2 (29) | 5 (71) |
| Difficult digestion | — | — | — | 3 (100) | — | 1 (14) | — | — | — | 1 (14) |
| Intestinal dilatation | 1 (33) | — | — | — | — | 1 (14) | 1 (14) | — | — | 1 (14) |
| Myochosis | — | 2 (67) | 1 (33) | — | — | — | 1 (14) | — | 1 (14) | 3 (43) |
| Non-erosive reflux disease | 3 (100) | — | — | — | — | 1 (14) | — | — | 3 (43) | 3 (43) |
| Pancreatic enzyme abnormality | — | — | 1 (33) | 1 (33) | 1 (33) | 2 (29) | 1 (14) | 1 (14) | 1 (14) | — |
Modified
tbl2 %>%
modify_header(all_ae_cols() ~ "**Grade {by}**") %>%
modify_spanning_header(all_ae_cols() ~ "**{strata}** \n{style_percent(p)}% ({n}/{N})") %>%
modify_caption("My caption: N = {N}") %>%
modify_footnote(label = "My footnote: N = {N}") | Adverse Event1 |
Drug A
30% (3/10) |
Drug B
70% (7/10) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | |
| Blood and lymphatic system disorders | — | 1 (33) | — | 1 (33) | 1 (33) | — | — | — | 1 (14) | 6 (86) |
| Anaemia | — | — | 1 (33) | 1 (33) | — | — | — | 1 (14) | 1 (14) | 3 (43) |
| Increased tendency to bruise | — | — | — | 1 (33) | — | — | — | — | 3 (43) | 2 (29) |
| Iron deficiency anaemia | — | — | — | 1 (33) | 1 (33) | 1 (14) | 2 (29) | — | 1 (14) | 1 (14) |
| Thrombocytopenia | — | 1 (33) | — | 1 (33) | — | — | — | 3 (43) | — | 4 (57) |
| Gastrointestinal disorders | — | — | — | 2 (67) | 1 (33) | — | — | — | 2 (29) | 5 (71) |
| Difficult digestion | — | — | — | 3 (100) | — | 1 (14) | — | — | — | 1 (14) |
| Intestinal dilatation | 1 (33) | — | — | — | — | 1 (14) | 1 (14) | — | — | 1 (14) |
| Myochosis | — | 2 (67) | 1 (33) | — | — | — | 1 (14) | — | 1 (14) | 3 (43) |
| Non-erosive reflux disease | 3 (100) | — | — | — | — | 1 (14) | — | — | 3 (43) | 3 (43) |
| Pancreatic enzyme abnormality | — | — | 1 (33) | 1 (33) | 1 (33) | 2 (29) | 1 (14) | 1 (14) | 1 (14) | — |
| 1 My footnote: N = 10 | ||||||||||
tbl2 %>%
modify_spanning_header(
all_cols_in_strata("Drug A") ~ "**Control Group** \n{style_percent(p)}% ({n}/{N})",
all_cols_in_strata("Drug B") ~ "**Experimental Group** \n{style_percent(p)}% ({n}/{N})"
)| Adverse Event |
Control Group
30% (3/10) |
Experimental Group
70% (7/10) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Blood and lymphatic system disorders | — | 1 (33) | — | 1 (33) | 1 (33) | — | — | — | 1 (14) | 6 (86) |
| Anaemia | — | — | 1 (33) | 1 (33) | — | — | — | 1 (14) | 1 (14) | 3 (43) |
| Increased tendency to bruise | — | — | — | 1 (33) | — | — | — | — | 3 (43) | 2 (29) |
| Iron deficiency anaemia | — | — | — | 1 (33) | 1 (33) | 1 (14) | 2 (29) | — | 1 (14) | 1 (14) |
| Thrombocytopenia | — | 1 (33) | — | 1 (33) | — | — | — | 3 (43) | — | 4 (57) |
| Gastrointestinal disorders | — | — | — | 2 (67) | 1 (33) | — | — | — | 2 (29) | 5 (71) |
| Difficult digestion | — | — | — | 3 (100) | — | 1 (14) | — | — | — | 1 (14) |
| Intestinal dilatation | 1 (33) | — | — | — | — | 1 (14) | 1 (14) | — | — | 1 (14) |
| Myochosis | — | 2 (67) | 1 (33) | — | — | — | 1 (14) | — | 1 (14) | 3 (43) |
| Non-erosive reflux disease | 3 (100) | — | — | — | — | 1 (14) | — | — | 3 (43) | 3 (43) |
| Pancreatic enzyme abnormality | — | — | 1 (33) | 1 (33) | 1 (33) | 2 (29) | 1 (14) | 1 (14) | 1 (14) | — |
Summary tables
For tbl_reg_summary(), the modify_header()
and modify_spanning_header() functions work with the
{gtsummary} column selectors like all_stat_cols() to
specify formatting of table headers.
tbl_reg_summary(), without overall
tbl3 <-
df_patient_characteristics %>%
select(trt, marker, discontinued) %>%
tbl_reg_summary(
by = trt
) %>%
bold_labels()Unmodified
# show_header_names(tbl3)
tbl3| Characteristic |
Drug A N = 441 |
Drug B N = 561 |
|---|---|---|
| Biological Marker | ||
| N Non-missing | 44 | 56 |
| Mean (SD) | 5.47 (0.97) | 5.46 (1.04) |
| Median (Q1, Q3) | 5.27 (4.96, 5.94) | 5.43 (4.65, 6.11) |
| Min, Max | 3.53, 8.42 | 3.66, 8.00 |
| N Missing | 0 | 0 |
| Discontinued from Study | 17 (39%) | 23 (41%) |
| 1 n (%) | ||
Modified
tbl3 %>%
modify_header(
all_stat_cols() ~ "**{level}**, N = {n}/{N} ({style_percent(p)}%)"
)| Characteristic | Drug A, N = 44/100 (44%)1 | Drug B, N = 56/100 (56%)1 |
|---|---|---|
| Biological Marker | ||
| N Non-missing | 44 | 56 |
| Mean (SD) | 5.47 (0.97) | 5.46 (1.04) |
| Median (Q1, Q3) | 5.27 (4.96, 5.94) | 5.43 (4.65, 6.11) |
| Min, Max | 3.53, 8.42 | 3.66, 8.00 |
| N Missing | 0 | 0 |
| Discontinued from Study | 17 (39%) | 23 (41%) |
| 1 n (%) | ||
tbl_reg_summary(), with overall
tbl4 <-
df_patient_characteristics %>%
select(trt, marker, discontinued) %>%
tbl_reg_summary(
by = trt
) %>%
add_overall(last = TRUE) %>%
bold_labels()Unmodified
# show_header_names(tbl4)
tbl4| Characteristic |
Drug A N = 441 |
Drug B N = 561 |
Overall N = 1001 |
|---|---|---|---|
| Biological Marker | |||
| N Non-missing | 44 | 56 | 100 |
| Mean (SD) | 5.47 (0.97) | 5.46 (1.04) | 5.47 (1.00) |
| Median (Q1, Q3) | 5.27 (4.96, 5.94) | 5.43 (4.65, 6.11) | 5.37 (4.76, 6.05) |
| Min, Max | 3.53, 8.42 | 3.66, 8.00 | 3.53, 8.42 |
| N Missing | 0 | 0 | 0 |
| Discontinued from Study | 17 (39%) | 23 (41%) | 40 (40%) |
| 1 n (%) | |||
Modified
tbl4 %>%
modify_header(
list(
stat_1 ~ "**Control Group** \n N = {n}",
stat_2 ~ "**Experimental Group** \n N = {n}",
stat_0 ~ "**Overall** \n N = {N}"
))
#> Warning: The `update` argument of `modify_header()` is deprecated as of gtsummary 2.0.0.
#> ℹ Use `modify_header(...)` input instead. Dynamic dots allow for syntax like
#> `modify_header(!!!list(...))`.
#> ℹ The deprecated feature was likely used in the gtsummary package.
#> Please report the issue at <https://github.com/ddsjoberg/gtsummary/issues>.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.| Characteristic |
Control Group N = 441 |
Experimental Group N = 561 |
Overall N = 1001 |
|---|---|---|---|
| Biological Marker | |||
| N Non-missing | 44 | 56 | 100 |
| Mean (SD) | 5.47 (0.97) | 5.46 (1.04) | 5.47 (1.00) |
| Median (Q1, Q3) | 5.27 (4.96, 5.94) | 5.43 (4.65, 6.11) | 5.37 (4.76, 6.05) |
| Min, Max | 3.53, 8.42 | 3.66, 8.00 | 3.53, 8.42 |
| N Missing | 0 | 0 | 0 |
| Discontinued from Study | 17 (39%) | 23 (41%) | 40 (40%) |
| 1 n (%) | |||