library(tidyverse) # general use
library(janitor) # handy data cleaning functions
library(readxl) # import excel files
library(here) # find files in a project
library(gtsummary) # presentation ready summary tables Data Cleaning: Exercises
Instructions
Recommended: Complete these exercises in the dedicated Posit Cloud work space, which comes with
all packages pre-installed, and
a Quarto document to fill in.
It will be helpful to peek here to verify that your tables generated in Posit Cloud match the desired output and code solutions.
Join R/Medicine Posit cloud work space for Cleaning Medical Data with R: bit.ly/posit-cloud-cleaning-medical-data
Otherwise: Follow along this document, work on your personal computer, and challenge yourself not to peek at the code solutions until you have completed the exercise.
Packages
The Data
The exercises use messy_uc.xlsx.
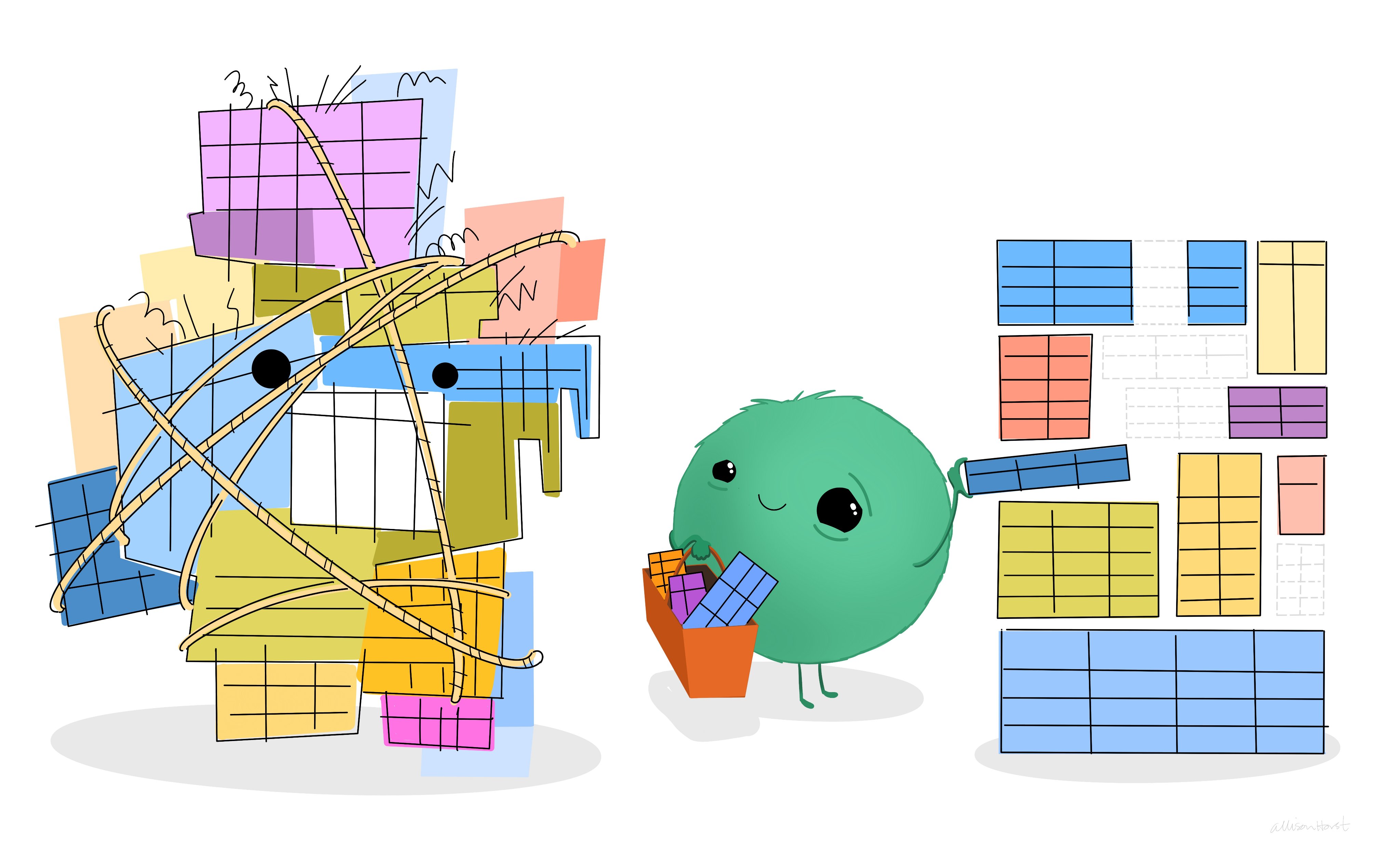
The data for each section will start out as messy Excel files of the type your well-meaning clinical colleague will have used to collect data from the local electronic medical record (EMR), with typos, data problems, and often untidy data structures.
Essentially, your job is to turn the messy Excel data beast (on the left) into the tidy data on the right.
 CL: Principles of Data Management
CL: Principles of Data Management
CL1
- Import
messy_uc.xlsxusingreadxl::read_excel().
# import raw data
df_raw <- read_excel(
path = "data/messy_uc.xlsx",
sheet = "__",
skip = __
)Code
# import raw data
df_raw <- read_excel(
path = "data/messy_uc.xlsx",
sheet = "Data",
skip = 6
)CL2
- Use an exploratory function to review the data.
Code
dplyr::glimpse(df_raw)
skimr::skim(df_raw)
base::summary(df_raw)
visdat::vis_dat(df_raw)
summarytools::view(summarytools::dfSummary(df_raw))
DataExplorer::create_report(df_raw)
Hmisc::describe(df_raw) SP: Stage 1 Data Cleaning
SP: Stage 1 Data Cleaning
Set up
# do initial cleaning of variable names and removing empty rows/columns
df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols"))SP1
- Explore the values of
race.
Code
df_clean |> count(race)# A tibble: 9 × 2
race n
<chr> <int>
1 African-American 4
2 AmerInd 1
3 Asian 1
4 Caucasian 17
5 H/API 1
6 Hawaiian 1
7 Mixed 1
8 Other 2
9 afromerican 2- In the
df_cleandata set, create a new variable namedrace_cleanthat cleans the coding ofrace(combine “African-American” & “afromerican”; “H/API” & “Mixed” & “Other”).
Code
df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
race_clean = case_when(
race %in% c("African-American", "afromerican") ~ "African-American",
race %in% c("H/API", "Mixed", "Other") ~ "Other",
.default = race
)
)- Confirm the new
race_cleanvariable is coded correctly.
Code
df_clean |>
count(race_clean, race)# A tibble: 9 × 3
race_clean race n
<chr> <chr> <int>
1 African-American African-American 4
2 African-American afromerican 2
3 AmerInd AmerInd 1
4 Asian Asian 1
5 Caucasian Caucasian 17
6 Hawaiian Hawaiian 1
7 Other H/API 1
8 Other Mixed 1
9 Other Other 2SP2
- Explore the type of and values of
start_plt.
Code
df_clean |>
count(start_plt)# A tibble: 28 × 2
start_plt n
<chr> <int>
1 115K/microL 1
2 1550K/microL 1
3 177K/microL 1
4 188K/microL 1
5 197K/microL 1
6 204K/microL 1
7 249K/microL 1
8 258K/microL 1
9 273K/microL 1
10 288K/microL 1
# ℹ 18 more rowsCode
df_clean |>
select(start_plt) |>
glimpse()Rows: 30
Columns: 1
$ start_plt <chr> "273K/microL", "414K/microL", "323K/microL", "389K/microL", …Code
df_clean[["start_plt"]] [1] "273K/microL" "414K/microL" "323K/microL" "389K/microL" "411K/microL"
[6] "427K/microL" "249K/microL" "197K/microL" "204K/microL" "305K/microL"
[11] "347K/microL" "402K/microL" "389K/microL" "432K/microL" "288K/microL"
[16] "177K/microL" "290K/microL" "312K/microL" "399K/microL" "423K/microL"
[21] "clumped" "323K/microL" "258K/microL" "115K/microL" "1550K/microL"
[26] "37K/microL" "188K/microL" "456K/microL" "356K/microL" "291K/microL" - In the
df_cleandata set, create a new variable namedstart_plt_cleanthat corrects any unusual values and assigns the correct variable type.
Code
df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
race_clean = case_when(
race %in% c("African-American", "afromerican") ~ "African-American",
race %in% c("H/API", "Mixed", "Other") ~ "Other",
.default = race
),
start_plt_clean = parse_number(start_plt, na = "clumped")
)- Confirm the new
start_plt_cleanvariable is coded correctly.
Code
df_clean |>
count(start_plt_clean, start_plt)# A tibble: 28 × 3
start_plt_clean start_plt n
<dbl> <chr> <int>
1 37 37K/microL 1
2 115 115K/microL 1
3 177 177K/microL 1
4 188 188K/microL 1
5 197 197K/microL 1
6 204 204K/microL 1
7 249 249K/microL 1
8 258 258K/microL 1
9 273 273K/microL 1
10 288 288K/microL 1
# ℹ 18 more rowsCode
df_clean |>
select(start_plt, start_plt_clean) |>
glimpse()Rows: 30
Columns: 2
$ start_plt <chr> "273K/microL", "414K/microL", "323K/microL", "389K/mic…
$ start_plt_clean <dbl> 273, 414, 323, 389, 411, 427, 249, 197, 204, 305, 347,…Code
df_clean[["start_plt_clean"]] [1] 273 414 323 389 411 427 249 197 204 305 347 402 389 432 288
[16] 177 290 312 399 423 NA 323 258 115 1550 37 188 456 356 291SP3
- Explore the type of and values of
race_clean.
Code
df_clean |>
count(race_clean)# A tibble: 6 × 2
race_clean n
<chr> <int>
1 African-American 6
2 AmerInd 1
3 Asian 1
4 Caucasian 17
5 Hawaiian 1
6 Other 4- Convert the
race_cleanvariable to a factor such that the most common values present in order in a summary table.
Code
df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
race_clean = case_when(
race %in% c("African-American", "afromerican") ~ "African-American",
race %in% c("H/API", "Mixed", "Other") ~ "Other",
.default = race
) |> fct_infreq(),
start_plt_clean = parse_number(start_plt)
)- Confirm the new coding of
race_clean.
Code
df_clean |>
count(race_clean, race)# A tibble: 9 × 3
race_clean race n
<fct> <chr> <int>
1 Caucasian Caucasian 17
2 African-American African-American 4
3 African-American afromerican 2
4 Other H/API 1
5 Other Mixed 1
6 Other Other 2
7 AmerInd AmerInd 1
8 Asian Asian 1
9 Hawaiian Hawaiian 1Code
df_clean |>
count(race_clean)# A tibble: 6 × 2
race_clean n
<fct> <int>
1 Caucasian 17
2 African-American 6
3 Other 4
4 AmerInd 1
5 Asian 1
6 Hawaiian 1 PH: Stage 2 Data Cleaning
PH: Stage 2 Data Cleaning
PH1
Pivoting Longer
Your Turn with endo_data
Measurements of Trans-Epithelial Electrical Resistance (TEER, the inverse of leakiness) in biopsies of 3 segments of intestine.
This could be affected by portal hypertension in patients with liver cirrhosis
Let’s find out!
Here is the code to load the data if you are doing this on a local computer. Use the clipboard icon at the top right to copy the code.
endo_data <- tibble::tribble(
~pat_id, ~portal_htn, ~duod_teer, ~ileal_teer, ~colon_teer,
001, 1, 4.33, 14.57, 16.23,
002, 0, 11.67, 15.99, 18.97,
003, 1, 4.12, 13.77, 15.22,
004, 1, 4.62, 16.37, 18.12,
005, 0, 12.43, 15.84, 19.04,
006, 0, 13.05, 16.23, 18.81,
007, 0, 11.88, 15.72, 18.31,
008, 1, 4.87, 16.59, 18.77,
009, 1, 4.23, 15.04, 16.87,
010, 0, 12.77, 16.73, 19.12
)
endo_data# A tibble: 10 × 5
pat_id portal_htn duod_teer ileal_teer colon_teer
<dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 4.33 14.6 16.2
2 2 0 11.7 16.0 19.0
3 3 1 4.12 13.8 15.2
4 4 1 4.62 16.4 18.1
5 5 0 12.4 15.8 19.0
6 6 0 13.0 16.2 18.8
7 7 0 11.9 15.7 18.3
8 8 1 4.87 16.6 18.8
9 9 1 4.23 15.0 16.9
10 10 0 12.8 16.7 19.1Pivoting Longer with endo_data
# A tibble: 10 × 5
pat_id portal_htn duod_teer ileal_teer colon_teer
<dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 4.33 14.6 16.2
2 2 0 11.7 16.0 19.0
3 3 1 4.12 13.8 15.2
4 4 1 4.62 16.4 18.1
5 5 0 12.4 15.8 19.0
6 6 0 13.0 16.2 18.8
7 7 0 11.9 15.7 18.3
8 8 1 4.87 16.6 18.8
9 9 1 4.23 15.0 16.9
10 10 0 12.8 16.7 19.1- What values do you want to use for these arguments to
pivot_longer:- cols
- names_pattern = “(.+)_teer”
- names_to
- values_to
- Note that we are giving you the correct value for
names_pattern, which will ask for what we want - to keep the characters of the name (of whatever length) before “_teer”
- Fill in the blanks to pivot this dataset to tall format, with columns for the intestinal location and the teer value.
- Note that we are giving you the correct answer for the
names_patternargument.
endo_data |>
pivot_longer(
cols = ___ ,
names_pattern = "(.+)_teer",
names_to = ___ ,
values_to = ___
)- Fill in the blanks to pivot this dataset to tall format, with columns for the intestinal location and the teer value.
Code
endo_data |>
pivot_longer(
cols = "duod_teer":"colon_teer",
names_pattern = "(.+)_teer",
names_to = c("location"),
values_to = "teer"
)- Run the code, and look at the resulting table. Use the clipboard icon at the top right to copy the code.
# A tibble: 30 × 4
pat_id portal_htn location teer
<dbl> <dbl> <chr> <dbl>
1 1 1 duod 4.33
2 1 1 ileal 14.6
3 1 1 colon 16.2
4 2 0 duod 11.7
5 2 0 ileal 16.0
6 2 0 colon 19.0
7 3 1 duod 4.12
8 3 1 ileal 13.8
9 3 1 colon 15.2
10 4 1 duod 4.62
# ℹ 20 more rows- Do you think that portal hypertension has an effect on TEER and (its inverse) epithelial leakiness?
PH2
Patient Demographics with Lab results (Your Turn to Join)
- We have some basic Patient Demographics in one table
# A tibble: 9 × 3
pat_id name age
<chr> <chr> <dbl>
1 001 Arthur Blankenship 67
2 002 Britney Jonas 23
3 003 Sally Davis 63
4 004 Al Jones 44
5 005 Gary Hamill 38
6 006 Ken Bartoletti 33
7 007 Ike Gerhold 52
8 008 Tatiana Grant 42
9 009 Antione Delacroix 27and potassium levels and creatinine levels in 2 other tables
# A tibble: 6 × 2
pat_id k
<chr> <dbl>
1 001 3.2
2 002 3.7
3 003 4.2
4 004 4.4
5 005 4.1
6 006 4 # A tibble: 6 × 2
pat_id cr
<chr> <dbl>
1 001 0.2
2 002 0.5
3 003 0.9
4 004 1.5
5 005 0.7
6 006 0.9Need to Load the Data?
If you are trying this on your local computer, copy the code below with the clipboard icon to get the data into your computer.
Code
demo <- tibble::tribble(
~pat_id, ~name, ~age,
'001', "Arthur Blankenship", 67,
'002', "Britney Jonas", 23,
'003', "Sally Davis", 63,
'004', "Al Jones", 44,
'005', "Gary Hamill", 38,
'006', "Ken Bartoletti", 33,
'007', "Ike Gerhold", 52,
'008', "Tatiana Grant", 42,
'009', "Antoine Delacroix", 27,
)
pot <- tibble::tribble(
~pat_id, ~k,
'001', 3.2,
'002', 3.7,
'003', 4.2,
'004', 4.4,
'005', 4.1,
'006', 4.0,
'007', 3.6,
'008', 4.2,
'009', 4.9,
)
cr <- tibble::tribble(
~pat_id, ~cr,
'001', 0.2,
'002', 0.5,
'003', 0.9,
'004', 1.5,
'005', 0.7,
'006', 0.9,
'007', 0.7,
'008', 1.0,
'009', 1.7,
)Your Turn to Join
- We want to join the correct labs (9 rows each) to the correct patients.
- The unique identifier (called the uniqid or key or recordID) is pat_id.
- It only occurs once for each patient/row
- It appears in each table we want to join
- The pat_id is of the character type in each (a common downfall if one is character, one is numeric, but they can look the same - but don’t match)
- We want to start with demographics, then add datasets that match to the right.
- We will use demo as our base dataset on the left hand side (LHS), and first join the potassium (pot) results (RHS)
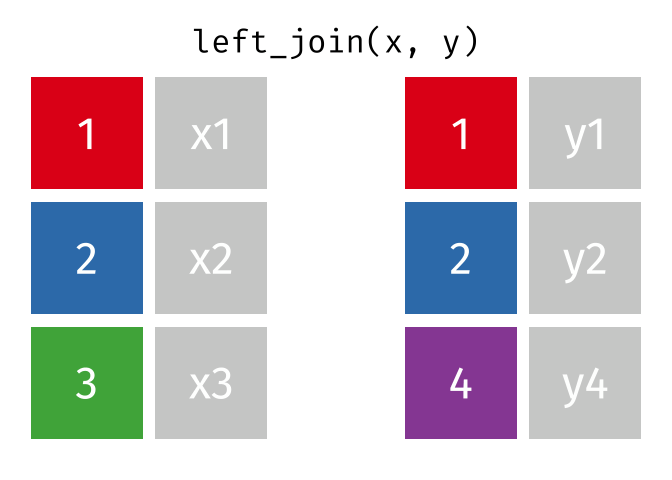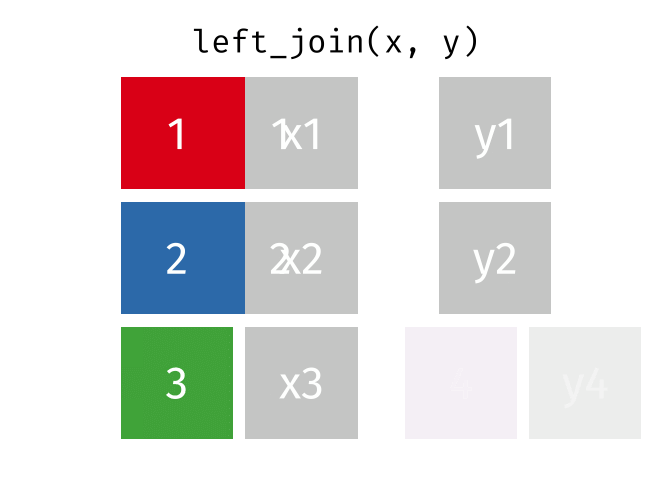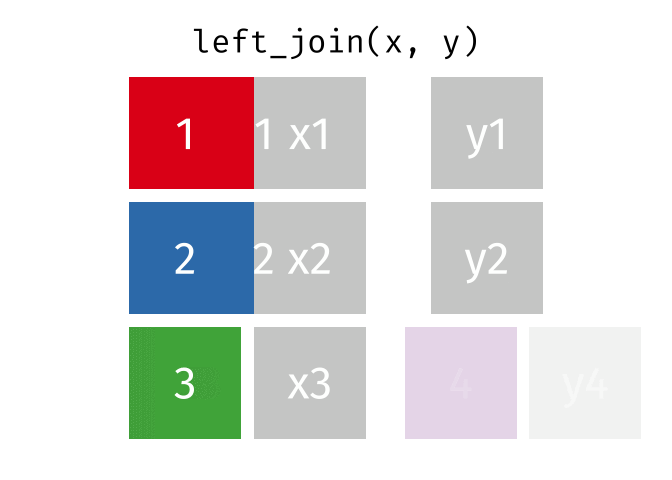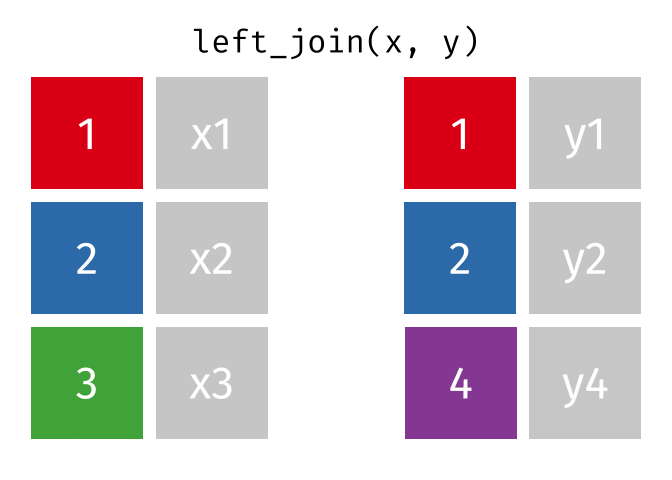
What the Left Join Looks Like
Your Turn to Join
- Joining demo to pot with a left_join
- left_join(data_x, data_y, by = “uniqid”)
- replace the generic arguments below with the correct ones to join demo to pot and produce new_data.
new_data <- left_join(data_x, data_y, by = "uniqid")
new_dataCode
new_data <- left_join(demo, pot, by = "pat_id")
new_data# A tibble: 9 × 4
pat_id name age k
<chr> <chr> <dbl> <dbl>
1 001 Arthur Blankenship 67 3.2
2 002 Britney Jonas 23 3.7
3 003 Sally Davis 63 4.2
4 004 Al Jones 44 4.4
5 005 Gary Hamill 38 4.1
6 006 Ken Bartoletti 33 4
7 007 Ike Gerhold 52 3.6
8 008 Tatiana Grant 42 4.2
9 009 Antoine Delacroix 27 4.9Now add Creatinine (cr) to new_data
- Joining new_data and cr with a left_join
- left_join(data_x, data_y, by = “uniqid”)
- Replace the generic arguments with the correct ones to join new_data and cr and produce new_data2.
new_data2 <- left_join(data_x, data_y, by = "uniqid")
new_data2Code
new_data2 <- left_join(new_data, cr, by = "pat_id")
new_data2# A tibble: 9 × 5
pat_id name age k cr
<chr> <chr> <dbl> <dbl> <dbl>
1 001 Arthur Blankenship 67 3.2 0.2
2 002 Britney Jonas 23 3.7 0.5
3 003 Sally Davis 63 4.2 0.9
4 004 Al Jones 44 4.4 1.5
5 005 Gary Hamill 38 4.1 0.7
6 006 Ken Bartoletti 33 4 0.9
7 007 Ike Gerhold 52 3.6 0.7
8 008 Tatiana Grant 42 4.2 1
9 009 Antoine Delacroix 27 4.9 1.7- Al has HTN and DM2
- Antoine has early stage FSGS
Session info
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.0 (2023-04-21 ucrt)
os Windows 10 x64 (build 19045)
system x86_64, mingw32
ui RTerm
language (EN)
collate English_United States.utf8
ctype English_United States.utf8
tz America/New_York
date 2024-01-02
pandoc 3.1.1 @ C:/Program Files/RStudio/resources/app/bin/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
! package * version date (UTC) lib source
P broom.helpers 1.13.0 2023-03-28 [?] CRAN (R 4.3.0)
P cachem 1.0.8 2023-05-01 [?] CRAN (R 4.3.0)
P callr 3.7.3 2022-11-02 [?] CRAN (R 4.3.0)
P cellranger 1.1.0 2016-07-27 [?] CRAN (R 4.3.0)
P cli 3.6.1 2023-03-23 [?] CRAN (R 4.3.0)
P colorspace 2.1-0 2023-01-23 [?] CRAN (R 4.3.0)
P crayon 1.5.2 2022-09-29 [?] CRAN (R 4.3.0)
P devtools 2.4.5 2022-10-11 [?] CRAN (R 4.3.0)
P digest 0.6.31 2022-12-11 [?] CRAN (R 4.3.0)
P dplyr * 1.1.2 2023-04-20 [?] CRAN (R 4.3.0)
P ellipsis 0.3.2 2021-04-29 [?] CRAN (R 4.3.0)
P evaluate 0.21 2023-05-05 [?] CRAN (R 4.3.0)
P fansi 1.0.4 2023-01-22 [?] CRAN (R 4.3.0)
P fastmap 1.1.1 2023-02-24 [?] CRAN (R 4.3.0)
P forcats * 1.0.0 2023-01-29 [?] CRAN (R 4.3.0)
P fs 1.6.2 2023-04-25 [?] CRAN (R 4.3.0)
P generics 0.1.3 2022-07-05 [?] CRAN (R 4.3.0)
P ggplot2 * 3.4.2 2023-04-03 [?] CRAN (R 4.3.0)
P glue 1.6.2 2022-02-24 [?] CRAN (R 4.3.0)
P gt 0.9.0 2023-03-31 [?] CRAN (R 4.3.0)
P gtable 0.3.3 2023-03-21 [?] CRAN (R 4.3.0)
P gtsummary * 1.7.1 2023-04-27 [?] CRAN (R 4.3.0)
P here * 1.0.1 2020-12-13 [?] CRAN (R 4.3.0)
P hms 1.1.3 2023-03-21 [?] CRAN (R 4.3.0)
P htmltools 0.5.5 2023-03-23 [?] CRAN (R 4.3.0)
P htmlwidgets 1.6.2 2023-03-17 [?] CRAN (R 4.3.0)
P httpuv 1.6.11 2023-05-11 [?] CRAN (R 4.3.0)
P janitor * 2.2.0 2023-02-02 [?] CRAN (R 4.3.0)
P jsonlite 1.8.4 2022-12-06 [?] CRAN (R 4.3.0)
P knitr 1.42 2023-01-25 [?] CRAN (R 4.3.0)
P later 1.3.1 2023-05-02 [?] CRAN (R 4.3.0)
P lifecycle 1.0.3 2022-10-07 [?] CRAN (R 4.3.0)
P lubridate * 1.9.2 2023-02-10 [?] CRAN (R 4.3.0)
P magrittr 2.0.3 2022-03-30 [?] CRAN (R 4.3.0)
P memoise 2.0.1 2021-11-26 [?] CRAN (R 4.3.0)
P mime 0.12 2021-09-28 [?] CRAN (R 4.3.0)
P miniUI 0.1.1.1 2018-05-18 [?] CRAN (R 4.3.0)
P munsell 0.5.0 2018-06-12 [?] CRAN (R 4.3.0)
P pillar 1.9.0 2023-03-22 [?] CRAN (R 4.3.0)
P pkgbuild 1.4.0 2022-11-27 [?] CRAN (R 4.3.0)
P pkgconfig 2.0.3 2019-09-22 [?] CRAN (R 4.3.0)
P pkgload 1.3.2 2022-11-16 [?] CRAN (R 4.3.0)
P prettyunits 1.1.1 2020-01-24 [?] CRAN (R 4.3.0)
P processx 3.8.1 2023-04-18 [?] CRAN (R 4.3.0)
P profvis 0.3.8 2023-05-02 [?] CRAN (R 4.3.0)
P promises 1.2.0.1 2021-02-11 [?] CRAN (R 4.3.0)
P ps 1.7.5 2023-04-18 [?] CRAN (R 4.3.0)
P purrr * 1.0.1 2023-01-10 [?] CRAN (R 4.3.0)
P R6 2.5.1 2021-08-19 [?] CRAN (R 4.3.0)
P Rcpp 1.0.10 2023-01-22 [?] CRAN (R 4.3.0)
P readr * 2.1.4 2023-02-10 [?] CRAN (R 4.3.0)
P readxl * 1.4.2 2023-02-09 [?] CRAN (R 4.3.0)
P remotes 2.4.2 2021-11-30 [?] CRAN (R 4.3.0)
renv 0.17.2 2023-03-17 [1] CRAN (R 4.3.0)
P rlang 1.1.1 2023-04-28 [?] CRAN (R 4.3.0)
P rmarkdown 2.21 2023-03-26 [?] CRAN (R 4.3.0)
P rprojroot 2.0.3 2022-04-02 [?] CRAN (R 4.3.0)
P rstudioapi 0.14 2022-08-22 [?] CRAN (R 4.3.0)
P scales 1.2.1 2022-08-20 [?] CRAN (R 4.3.0)
P sessioninfo 1.2.2 2021-12-06 [?] CRAN (R 4.3.0)
P shiny 1.7.4 2022-12-15 [?] CRAN (R 4.3.0)
P snakecase 0.11.0 2019-05-25 [?] CRAN (R 4.3.0)
P stringi 1.7.12 2023-01-11 [?] CRAN (R 4.3.0)
P stringr * 1.5.0 2022-12-02 [?] CRAN (R 4.3.0)
P tibble * 3.2.1 2023-03-20 [?] CRAN (R 4.3.0)
P tidyr * 1.3.0 2023-01-24 [?] CRAN (R 4.3.0)
P tidyselect 1.2.0 2022-10-10 [?] CRAN (R 4.3.0)
P tidyverse * 2.0.0 2023-02-22 [?] CRAN (R 4.3.0)
P timechange 0.2.0 2023-01-11 [?] CRAN (R 4.3.0)
P tzdb 0.4.0 2023-05-12 [?] CRAN (R 4.3.0)
P urlchecker 1.0.1 2021-11-30 [?] CRAN (R 4.3.0)
P usethis 2.1.6 2022-05-25 [?] CRAN (R 4.3.0)
P utf8 1.2.3 2023-01-31 [?] CRAN (R 4.3.0)
P vctrs 0.6.2 2023-04-19 [?] CRAN (R 4.3.0)
P withr 2.5.0 2022-03-03 [?] CRAN (R 4.3.0)
P xfun 0.39 2023-04-20 [?] CRAN (R 4.3.0)
P xml2 1.3.4 2023-04-27 [?] CRAN (R 4.3.0)
P xtable 1.8-4 2019-04-21 [?] CRAN (R 4.3.0)
P yaml 2.3.7 2023-01-23 [?] CRAN (R 4.3.0)
[1] C:/Users/pileggis/Documents/gh-personal/rmedicine-data-cleaning-2023/renv/library/R-4.3/x86_64-w64-mingw32
[2] C:/Users/pileggis/AppData/Local/R/cache/R/renv/sandbox/R-4.3/x86_64-w64-mingw32/830ce55b
P ── Loaded and on-disk path mismatch.
──────────────────────────────────────────────────────────────────────────────