Cleaning Medical Data
with R
Introduction
Licensing
This work is licensed under Creative Commons 4.0 International.
Instructors
Crystal Lewis

Shannon Pileggi

Peter Higgins

Scope
Taming the Data Beast, by Allison Horst
Taming the Data Beast, from Allison Horst’s Data Science Illustrations
Schedule
| Time | Topic |
|---|---|
| 11:00 - 11:10 | Intro/Logistics |
| 11:10 - 12:00 | Crystal Lewis (Principles of Data Management) |
| 12:00 - 12:05 | Break |
| 12:05 - 12:55 | Shannon Pileggi (Stage 1 Data Cleaning) |
| 12:55 - 1:00 | Break |
| 01:00 - 02:00 | Peter Higgins (Stage 2 Data Cleaning) |
Please add any questions to the public Zoom chat. These may be answered in the moment or addressed at the end depending on context.
Exercises
All exercises can be accessed via Posit Cloud.
See exercise instructions for the link to access the Posit Cloud workspace.
Syntax aside
Pipes
2014+ magrittr pipe
%>%2021+ (R \(\geq\) 4.1.0) native R pipe
|>
2022 Isabella Velásquez Understanding the native R pipe |> https://ivelasq.rbind.io/blog/understanding-the-r-pipe/
R for Data Science: Ch 18 Pipes

Namespacing
package::function()
dplyr::select()
tells R explicitly to use the function
selectfrom the packagedplyrcan help to avoid name conflicts (e.g.,
MASS::select())does not require
library(dplyr)generally, we aimed to namespace functions from non-tidyverse packages.
Principles of Data Management
Data Horror Stories
Data Horror Stories

Image from knowyourmeme.com
Data Organizing Principles
- Data Structure
- Variable Values
- Variable Types
- Missing Data
Data Structure
- Data should make a rectangle of rows and columns
- You should have the expected number of rows (cases in your data)
- You should have the expected number of columns (variables in your data)
Data Structure
- Variable names should be the first, and only the first, row of your data
- They should also adhere to best practices
- Variable names should
- Be unique
- Be meaningful (
genderinstead ofX1) - Not include spaces
- Not include special characters except
_- So no
/,-,!,"
- So no
- Not start with a number or special character
Exercise
What data structure issues do you notice in our sample data?
01:00
Data Structure
- Variable names are not the first row of the data
- Our data does not make a rectangle - Empty column, empty rows
- Variable names do not adhere to best practices
Variable Values
- Values should be explicit, not implicit
- If a blank cell is implied to be zero, fill that cell with an actual zero
- No color coding should be used to indicate information. Make a new variable.
- Values should be analyzable. This means no more than one measure should be captured in a variable.
- Variables should be captured consistently within a column
- Dates captured consistently (i.e. YYYY-MM-DD)
- Categories captured consistently (both spelling and capitalization)
- If the variable is numeric, values should fall within your expected range
Exercise
What variable value issues do you notice in our sample data?
01:00
Variable Values
- Color coding used to indicate information
- Two things measured in one column
- Categorical values captured inconsistently
Variable Types
Variables should be stored as your expected type (or in R terms - class)
- Numeric
- Contain numeric values (14.5, 14.539, 789, -24)
- Numeric variables cannot contain special characters, spaces, or letters
- 100mg
- 83/150
- ” 89”
- Date, Time, Date-Time
- Represented in R as either
<date>,<time>or<dttm>/<POSIXct> - Allow you to perform calculations on dates
- Represented in R as either
Variable Types
- Character
- Contain character values or strings (“kg”, “R in Medicine”, “11.5”, “5mg”)
- Factor
- A special class of variables, helpful when working with categorical or ordinal variables
- Factors assign an order to your variable groups
- You must assign this class to your variables
- You can learn more about working with factors in this article: Wrangling Categorical Data in R
Exercise
What is the R class for the following variables?
Exercise
What variable type issues do you notice in our sample data?
01:00
Variable Types
- Dates stored as numbers
- Text stored in numeric variables
Missing Data
- Missing data should appear as you expect it to
- The amount of missingness
- The variables/cases that data is missing for
- Use consistent values to indicate missing responses (Blank, NA, -999)
- Document your decision
- The missing values should match your variable type
- i.e., Don’t use “no response” in a numeric variable
Exercise
What missing data issues do you notice in our sample data?
01:00
Missing Data
- Unexpected missing data
- Inconsistent missing values used
- Missing values do not match variable type
Error Reduction
The number one way to reduce data errors is to make a plan before you collect data
Correct data at the source
Plan the variables you want to collect
Build your data collection/entry tools in a way that follows your plan
Test your data tools before collecting/entering data
Check your data often during data collection/entry
Plan the variables you want to collect
Necessary to plan for
- Variable name
- Variable label/item wording
- Variable type
- Allowable values/ranges
- Missing values
Nice to plan for
- Skip patterns
- Required items
- Variable universe
Add those variables to a data dictionary
| var_name | label | type | values | missing_values |
|---|---|---|---|---|
| pat_id | Patient Identifier | character | 001-030 | NA |
| treatment | Treatment for UC | character | upa; uste; oza | NA |
| start_date | Date of start of treatment | date | YYYY-MM-DD | NA |
| ethnic | Ethnicity - hispanic or not hispanic | character | hispanic; not hispanic | missing |
| start_mes | Mayo endoscopic Score at start of treatment | numeric | 0-3 | -99 |
Build your tools based on your data dictionary
Excel
Qualtrics
REDCap
Know the strengths and limitations of your tool
- Consider things such a privacy (PHI), versioning, validation
- Tidy Spreadsheets in Medical Research (70 mins)
Build your tools based on your data dictionary
Name your variables correctly in your tool
- Instead of Q1, Q2, Q3 -> id, start_date, treatment
Build items to only accept allowable values
- Only within specified range (0-50)
- Only within specified categories (“hispanic”, “not hispanic”)
Build items to only accept specified variable types
- Only numeric values
- Only dates in the YYYY-MM-DD format
Test your data collection or entry tool
- Collect/enter sample data
- Are any items missing?
- Are you getting unexpected values for items?
- Values out of range
- Incorrect formats
- Inconsistent entries
- “m”, “male”, “Male”, “MALE”
- Is the skip logic working as expected?
- Are people able to skip items that they should not be able to skip?
Review your data often during data collection or entry
- Validate your data based on your expectations
pointblankvalidateassertrdataquieR- Excellent resource: Data Validation in R: From Principles to Tools and Packages (80 minutes)
pointblank report
library(pointblank)
# Import my data
df_raw <- readxl::read_excel("data/mydata.csv")
# Check my assumptions
create_agent(df_raw) |>
rows_distinct(columns = vars(pat_id)) |>
col_vals_not_null(columns = vars(pat_id)) |>
col_is_date(columns = vars(start_date)) |>
col_is_numeric(columns = vars(start_mes)) |>
col_vals_in_set(columns = vars(treatment), set = c("upa", "uste", "oza")) |>
col_vals_in_set(columns = vars(ethnic), set = c("hispanic", "not hispanic")) |>
col_vals_between(columns = vars(start_mes), left = 0, right = 3, na_pass = FALSE) |>
interrogate()Review your data often during data collection
- Create a codebook to review univariate summary statistics
codebookrcodebookmemiscsjPlot
codebookr codebook
Data Cleaning Practices
Scenario
We have data that originate from an observational study comparing 3 treatments of ulcerative colitis (UC)
We have an analysis question:
- Are there differences in change in MES and QOL scores between start and finish, and are the decreases in scores greater for any of the 3 new medications?
In order to answer this question, we have asked a student to extract data from the medical record into Excel
Along with the spreadsheet, we are provided a data dictionary
As we start to review the data, we find a sundry of errors that need correction
Exercise
Take 5 minutes to review the data dictionary and our data.
- Log in to Posit Cloud and navigate to our project
- Open the data folder and open the file “messy_uc.xlsx”
When you finish, give us a 👍
If you are having trouble, give us a ✋
05:00
Import our file
We are going to use the
read_excel()function from thereadxlpackageThere are several arguments to consider when using this function
- path
- sheet = NULL
- col_names = TRUE
- na = ” ”
- skip = 0
type
?read_excelin your console to see more arguments
Import our file
| pat_id | treatment | start_date | ethnic | race | dob | ...7 | start_bp | pre/post_wt_kg | start_mes |
|---|---|---|---|---|---|---|---|---|---|
| 001 | upa | 44208 | hispanic | Caucasian | 2005-01-07 | NA | 114/72 | 84/82 | 3 |
| 002 | uste | 44215 | not hispanic | Caucasian | 1937-04-13 | NA | 132/86 | 77/77 | 2 |
| 003 | oza | 44230 | not hispanic | African-American | 1946-06-06 | NA | 124/92 | 74/75 | 1 |
| 004 | upa | 44245 | not hispanic | Caucasian | 1963-07-14 | NA | 144/83 | 66/65 | 3 |
| 005 | oza | 44255 | not hispanic | Mixed | 1978-05-12 | NA | 122/78 | 55/56 | 3 |
| 006 | uste | 44259 | not hispanic | Other | 1992-04-03 | NA | 121/80 | 111/110 | 2 |
| 007 | uste | 44264 | not hispanic | Asian | 1955-08-22 | NA | 133/74 | 133/130 | 3 |
| 008 | oza | 44999 | Hispanic | afromerican | 1974-09-11 | NA | 116/73 | 74/76 | 3 |
| 009 | upa | 44276 | NOT hispanic | Caucasian | 1984-11-14 | NA | 118/66 | 82/80 | 2 |
| 010 | oza | 44278 | hispamnic | Caucasian | 1972-12-20 | NA | 122/78 | 85/87 | 3 |
Exercise (CL1)
Your turn! Take 3 minutes to import the data.
- Open “exercises.qmd” in our Posit Cloud project
- Navigate to ## CL1
- Update the code and run the code chunk using the green arrow
–> Take me to the exercises <–
03:00
Review the data
EDA is not a formal process with a strict set of rules. More than anything, EDA is a state of mind. During the initial phases of EDA you should feel free to investigate every idea that occurs to you. - R for Data Science
Image from giphy.com
Review the data
Get to know your data
- How many rows? How many columns?
- What are the variable types?
- What are variable values?
- How much missing data is there?
- How are variables related?
There are several functions that can be used to explore data
dplyr::glimpse()skimr::skim()base::summary()visdat:vis_dat()summarytools::dfSummary()DataExplorer::create_report()Hmisc::describe()
summarytools::dfSummary()
skimr::skim()
Exercise (CL2)
Use one or more of these exploratory packages to review your data. What fixes do you see that need to happen?
dplyr::glimpse()skimr::skim()base::summary()visdat:vis_dat()
summarytools::dfSummary()DataExplorer::create_report()Hmisc::describe()
–> Take me to the exercises <–
05:00
Stage 1
Data Cleaning
Variable names
Original variable names in excel:
Variable names import as shown, with modifications from readxl::read_excel() to ensure uniqueness:
Variable names, cleaner
Variable names as imported:
Remove empty columns or rows
# A tibble: 6 × 5
pat_id race dob x7 start_bp
<chr> <chr> <dttm> <lgl> <chr>
1 013 Caucasian 1948-02-27 00:00:00 NA 118/73
2 014 African-American 1966-04-22 00:00:00 NA 106/59
3 015 H/API 1978-08-11 00:00:00 NA 112/69
4 <NA> <NA> NA NA <NA>
5 016 African-American 1998-10-28 00:00:00 NA 114/76
6 017 Caucasian 2001-01-09 00:00:00 NA 124/80 # A tibble: 6 × 4
pat_id race dob start_bp
<chr> <chr> <dttm> <chr>
1 013 Caucasian 1948-02-27 00:00:00 118/73
2 014 African-American 1966-04-22 00:00:00 106/59
3 015 H/API 1978-08-11 00:00:00 112/69
4 016 African-American 1998-10-28 00:00:00 114/76
5 017 Caucasian 2001-01-09 00:00:00 124/80
6 018 Caucasian 1994-03-07 00:00:00 120/68 Rows: 31
Columns: 38
$ pat_id <chr> "001", "002", "003", "004", "005", "0…
$ treatment <chr> "upa", "uste", "oza", "upa", "oza", "…
$ start_date <dbl> 44208, 44215, 44230, 44245, 44255, 44…
$ ethnic <chr> "hispanic", "not hispanic", "not hisp…
$ race <chr> "Caucasian", "Caucasian", "African-Am…
$ dob <dttm> 2005-01-07, 1937-04-13, 1946-06-06, …
$ x7 <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ start_bp <chr> "114/72", "132/86", "124/92", "144/83…
$ pre_post_wt_kg <chr> "84/82", "77/77", "74/75", "66/65", "…
$ start_mes <dbl> 3, 2, 1, 3, 3, 2, 3, 3, 2, 3, 3, 3, 3…
$ start_bss <dbl> 75.85449, 54.65536, 28.46290, 77.2364…
$ start_abd_score <dbl> 76.80263, 53.56908, 30.82623, 77.7276…
$ start_sys <dbl> 81.63807, 53.39802, 29.50837, 79.0785…
$ start_coping <dbl> 50.75913, 28.23465, 20.03401, 45.1019…
$ start_emo <dbl> 79.31260, 58.55421, 32.22812, 75.6089…
$ daily_life_impact_score_at_start <dbl> 87.81723, 59.87844, 28.39321, 86.1173…
$ start_wbc <dbl> 8.2, 10.1, 5.5, 4.7, 8.9, 9.3, 5.6, 9…
$ start_plt <chr> "273K/microL", "414K/microL", "323K/m…
$ start_na <chr> "137mmol/L", "142mmol/L", "140mmol/L"…
$ start_k <chr> "3.7", "4.0999999999999996", "4.3", "…
$ end_month <dbl> 6, 6, 7, 7, 7, 8, 8, 8, 8, 8, 9, 9, 9…
$ end_day <dbl> 14, 21, 6, 22, 30, 4, 10, 15, 22, 26,…
$ end_year <dbl> 2021, 2021, 2021, 2021, 2021, 2021, 2…
$ end_mes <dbl> 0, 1, 1, 1, 2, 1, 2, 2, 0, 1, 0, 1, 1…
$ end_bss <dbl> 9.854493, 32.655357, 28.462897, 33.23…
$ end_abd <dbl> 16.802628, 33.569079, 30.826225, 37.7…
$ end_sys <dbl> 9.638072, 29.398023, 29.508373, 31.07…
$ end_coping <dbl> 23.759131, 19.234646, 20.034009, 27.1…
$ end_emo <chr> "39.760954572288782", "49.36928154791…
$ end_dl <chr> "9.4097102977463347", "35.23874059870…
$ end_wbc <dbl> 8.541698, 11.076921, 3.000000, 4.8581…
$ end_plt <dbl> 201, 340, 256, 327, 432, 348, 181, 12…
$ end_na <dbl> 137.3278, 142.2140, 140.0831, 139.158…
$ end_k <dbl> 3.958430, 4.174979, 4.433533, 3.53357…
$ fake_street <chr> "990 Mohammad Mountain", "8512 O'Conn…
$ fake_city <chr> "North Sigmundville", "Port Halstad",…
$ fake_state <chr> "New Mexico", "Missouri", "South Caro…
$ fake_zip <dbl> 96074, 11264, 57246, 31457, 30711, 52…Rows: 30
Columns: 37
$ pat_id <chr> "001", "002", "003", "004", "005", "0…
$ treatment <chr> "upa", "uste", "oza", "upa", "oza", "…
$ start_date <dbl> 44208, 44215, 44230, 44245, 44255, 44…
$ ethnic <chr> "hispanic", "not hispanic", "not hisp…
$ race <chr> "Caucasian", "Caucasian", "African-Am…
$ dob <dttm> 2005-01-07, 1937-04-13, 1946-06-06, …
$ start_bp <chr> "114/72", "132/86", "124/92", "144/83…
$ pre_post_wt_kg <chr> "84/82", "77/77", "74/75", "66/65", "…
$ start_mes <dbl> 3, 2, 1, 3, 3, 2, 3, 3, 2, 3, 3, 3, 3…
$ start_bss <dbl> 75.85449, 54.65536, 28.46290, 77.2364…
$ start_abd_score <dbl> 76.80263, 53.56908, 30.82623, 77.7276…
$ start_sys <dbl> 81.63807, 53.39802, 29.50837, 79.0785…
$ start_coping <dbl> 50.75913, 28.23465, 20.03401, 45.1019…
$ start_emo <dbl> 79.31260, 58.55421, 32.22812, 75.6089…
$ daily_life_impact_score_at_start <dbl> 87.81723, 59.87844, 28.39321, 86.1173…
$ start_wbc <dbl> 8.2, 10.1, 5.5, 4.7, 8.9, 9.3, 5.6, 9…
$ start_plt <chr> "273K/microL", "414K/microL", "323K/m…
$ start_na <chr> "137mmol/L", "142mmol/L", "140mmol/L"…
$ start_k <chr> "3.7", "4.0999999999999996", "4.3", "…
$ end_month <dbl> 6, 6, 7, 7, 7, 8, 8, 8, 8, 8, 9, 9, 9…
$ end_day <dbl> 14, 21, 6, 22, 30, 4, 10, 15, 22, 26,…
$ end_year <dbl> 2021, 2021, 2021, 2021, 2021, 2021, 2…
$ end_mes <dbl> 0, 1, 1, 1, 2, 1, 2, 2, 0, 1, 0, 1, 1…
$ end_bss <dbl> 9.854493, 32.655357, 28.462897, 33.23…
$ end_abd <dbl> 16.802628, 33.569079, 30.826225, 37.7…
$ end_sys <dbl> 9.638072, 29.398023, 29.508373, 31.07…
$ end_coping <dbl> 23.759131, 19.234646, 20.034009, 27.1…
$ end_emo <chr> "39.760954572288782", "49.36928154791…
$ end_dl <chr> "9.4097102977463347", "35.23874059870…
$ end_wbc <dbl> 8.541698, 11.076921, 3.000000, 4.8581…
$ end_plt <dbl> 201, 340, 256, 327, 432, 348, 181, 12…
$ end_na <dbl> 137.3278, 142.2140, 140.0831, 139.158…
$ end_k <dbl> 3.958430, 4.174979, 4.433533, 3.53357…
$ fake_street <chr> "990 Mohammad Mountain", "8512 O'Conn…
$ fake_city <chr> "North Sigmundville", "Port Halstad",…
$ fake_state <chr> "New Mexico", "Missouri", "South Caro…
$ fake_zip <dbl> 96074, 11264, 57246, 31457, 30711, 52…Recoding
df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
ethnic_clean = case_when(
ethnic %in% c("hispanic", "Hispanic", "hispamnic") ~ "hispanic",
ethnic %in% c("NOT hispanic", "not hispanic") ~ "not hispanic",
.default = ethnic
)
)
df_clean |>
count(ethnic_clean)# A tibble: 2 × 2
ethnic_clean n
<chr> <int>
1 hispanic 5
2 not hispanic 25Exercise
Complete Data Cleaning Fundamentals Exercise SP1.
–> Take me to the exercises <–
05:00
Replace values with missing
df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
ethnic_clean = case_when(
ethnic %in% c("hispanic", "Hispanic", "hispamnic") ~ "hispanic",
ethnic %in% c("NOT hispanic", "not hispanic") ~ "not hispanic",
),
end_na_clean = na_if(end_na, -99)
) Incorrect variable type
Rows: 31
Columns: 1
$ end_emo <chr> "39.760954572288782", "49.369281547911214", "35.90128630678054… [1] "39.760954572288782" "49.369281547911214" "35.901286306780541"
[4] "50.2057532077703" "64.973437247083666" "35.965982580017993"
[7] "57.76367623799068" "61.960515932643503" "27.768279853236116"
[10] "45.948161004250508" "37.34426764708568" "51.859745680890889"
[13] "55.055731509083827" "62.246053520477801" "not done"
[16] NA "51.794563346733348" "28.024233546743879"
[19] "58.368126398937022" "13.120426912160625" "45.417571554273906"
[22] "41.388894056374887" "27.007297504717883" "64.595741275696824"
[25] "50.243011921919624" "52.659962900705473" "41.581059324621933"
[28] "33.176949642484104" "67.258563644329342" "22.931081374117134"
[31] "31.118353038978491"df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
ethnic_clean = case_when(
ethnic %in% c("hispanic", "Hispanic", "hispamnic") ~ "hispanic",
ethnic %in% c("NOT hispanic", "not hispanic") ~ "not hispanic",
),
end_na_clean = na_if(end_na, -99),
end_emo_clean = na_if(end_emo, "not done") |> as.numeric()
) Rows: 30
Columns: 2
$ end_emo <chr> "39.760954572288782", "49.369281547911214", "35.90128630…
$ end_emo_clean <dbl> 39.76095, 49.36928, 35.90129, 50.20575, 64.97344, 35.965…# A tibble: 30 × 3
end_emo_clean end_emo n
<dbl> <chr> <int>
1 13.1 13.120426912160625 1
2 22.9 22.931081374117134 1
3 27.0 27.007297504717883 1
4 27.8 27.768279853236116 1
5 28.0 28.024233546743879 1
6 31.1 31.118353038978491 1
7 33.2 33.176949642484104 1
8 35.9 35.901286306780541 1
9 36.0 35.965982580017993 1
10 37.3 37.34426764708568 1
# ℹ 20 more rowsCorrecting dates
Rows: 31
Columns: 1
$ start_date <dbl> 44208, 44215, 44230, 44245, 44255, 44259, 44264, 44999, 442…df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
ethnic_clean = case_when(
ethnic %in% c("hispanic", "Hispanic", "hispamnic") ~ "hispanic",
ethnic %in% c("NOT hispanic", "not hispanic") ~ "not hispanic",
),
end_na_clean = na_if(end_na, -99),
end_emo_clean = na_if(end_emo, "not done") |> as.numeric(),
start_date_clean = janitor::convert_to_date(start_date)
) # A tibble: 30 × 3
start_date start_date_clean n
<dbl> <date> <int>
1 44208 2021-01-12 1
2 44215 2021-01-19 1
3 44230 2021-02-03 1
4 44245 2021-02-18 1
5 44255 2021-02-28 1
6 44259 2021-03-04 1
7 44264 2021-03-09 1
8 44276 2021-03-21 1
9 44278 2021-03-23 1
10 44297 2021-04-11 1
# ℹ 20 more rowsExtracting numbers from text
Rows: 31
Columns: 1
$ start_na <chr> "137mmol/L", "142mmol/L", "140mmol/L", "139mmol/L", "144mmol/… [1] "137mmol/L" "142mmol/L" "140mmol/L" "139mmol/L" "144mmol/L" "145mmol/L"
[7] "142mmol/L" "138mmol/L" "140mmol/L" "137mmol/L" "143mmol/L" "136mmol/L"
[13] "135mmol/L" "141mmol/L" "133mmol/L" NA "135mmol/L" "143mmol/L"
[19] "136mmol/L" "144mmol/L" "145mmol/L" "140mmol/L" "141mmol/L" "142mmol/L"
[25] "138mmol/L" "139mmol/L" "142mmol/L" "144mmol/L" "139mmol/L" "138mmol/L"
[31] "140mmol/L"df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
ethnic_clean = case_when(
ethnic %in% c("hispanic", "Hispanic", "hispamnic") ~ "hispanic",
ethnic %in% c("NOT hispanic", "not hispanic") ~ "not hispanic",
),
end_na_clean = na_if(end_na, -99),
end_emo_clean = na_if(end_emo, "not done") |> as.numeric(),
start_na_clean = parse_number(start_na)
) Rows: 30
Columns: 2
$ start_na <chr> "137mmol/L", "142mmol/L", "140mmol/L", "139mmol/L", "14…
$ start_na_clean <dbl> 137, 142, 140, 139, 144, 145, 142, 138, 140, 137, 143, …# A tibble: 12 × 3
start_na_clean start_na n
<dbl> <chr> <int>
1 133 133mmol/L 1
2 135 135mmol/L 2
3 136 136mmol/L 2
4 137 137mmol/L 2
5 138 138mmol/L 3
6 139 139mmol/L 3
7 140 140mmol/L 4
8 141 141mmol/L 2
9 142 142mmol/L 4
10 143 143mmol/L 2
11 144 144mmol/L 3
12 145 145mmol/L 2Exercise
Complete Data Cleaning Fundamentals Exercise SP2.
–> Take me to the exercises <–
05:00
Character variable should be a factor
Rows: 30
Columns: 2
$ treatment <chr> "upa", "uste", "oza", "upa", "oza", "uste", "uste", "oza"…
$ ethnic_clean <chr> "hispanic", "not hispanic", "not hispanic", "not hispanic…df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
ethnic_clean = case_when(
ethnic %in% c("hispanic", "Hispanic", "hispamnic") ~ "hispanic",
ethnic %in% c("NOT hispanic", "not hispanic") ~ "not hispanic",
) |> fct_infreq(),
end_na_clean = na_if(end_na, -99),
end_emo_clean = na_if(end_emo, "not done") |> as.numeric(),
start_na_clean = parse_number(start_na),
treatment = fct_relevel(treatment, "upa", "uste", "oza")
) See the forcats package for other factor handling solutions.
Rows: 30
Columns: 2
$ treatment <fct> upa, uste, oza, upa, oza, uste, uste, oza, upa, oza, upa,…
$ ethnic_clean <fct> hispanic, not hispanic, not hispanic, not hispanic, not h…Exercise
Complete Data Cleaning Fundamentals Exercise SP3.
–> Take me to the exercises <–
05:00
Separating values
Rows: 30
Columns: 1
$ start_bp <chr> "114/72", "132/86", "124/92", "144/83", "122/78", "121/80", "… [1] "114/72" "132/86" "124/92" "144/83" "122/78" "121/80" "133/74" "116/73"
[9] "118/66" "122/78" "126/82" "114/68" "118/73" "106/59" "112/69" "114/76"
[17] "124/80" "120/68" "119/77" "116/74" "121/80" "112/58" "117/67" "118/73"
[25] "116/74" "126/84" "144/96" "120/84" "115/75" "142/92"df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
ethnic_clean = case_when(
ethnic %in% c("hispanic", "Hispanic", "hispamnic") ~ "hispanic",
ethnic %in% c("NOT hispanic", "not hispanic") ~ "not hispanic",
) |> fct_infreq(),
end_na_clean = na_if(end_na, -99),
end_emo_clean = na_if(end_emo, "not done") |> as.numeric(),
start_na_clean = parse_number(start_na),
treatment = fct_relevel(treatment, "upa", "uste", "oza")
) |>
separate_wider_delim(start_bp, delim ="/", names = c("bp_systolic", "bp_diastolic"), cols_remove = FALSE) |>
mutate(across(c(bp_systolic, bp_diastolic), as.numeric)) Rows: 30
Columns: 3
$ start_bp <chr> "114/72", "132/86", "124/92", "144/83", "122/78", "121/80…
$ bp_systolic <dbl> 114, 132, 124, 144, 122, 121, 133, 116, 118, 122, 126, 11…
$ bp_diastolic <dbl> 72, 86, 92, 83, 78, 80, 74, 73, 66, 78, 82, 68, 73, 59, 6…Assigning labels
What does anything mean?
# first import data dictionary
df_dictionary <- read_excel(
path = here("data", "messy_uc.xlsx"),
sheet = "Data_Dictionary"
)
df_dictionary# A tibble: 33 × 5
Variable Details Units Range ...5
<chr> <chr> <chr> <chr> <chr>
1 pat_id Patient Identifier digi… 001-… <NA>
2 treatment Treatment for UC <NA> upad… <NA>
3 start_date Date of start of treatment digi… YYYY… <NA>
4 ethnic Ethnicity - hispanic or not hispanic <NA> hisp… <NA>
5 race Race - one of 7 choices <NA> cauc… <NA>
6 dob date of birth digi… YYYY… <NA>
7 start_bp blood pressure at start - systolic/diastolic mm Hg syst… <NA>
8 pre/post_wt_kg weight in kilograms at start/end kilo… 48-1… <NA>
9 start_mes Mayo endoscopic Score at start of treatment poin… 0-3 <NA>
10 start_bss Bowel symptom score at start poin… 0-100 QOL …
# ℹ 23 more rows# second create a named vector of variable names and variable labels
vec_variables <- df_dictionary |>
select(Variable, Details) |>
deframe()
vec_variables pat_id
"Patient Identifier"
treatment
"Treatment for UC"
start_date
"Date of start of treatment"
ethnic
"Ethnicity - hispanic or not hispanic"
race
"Race - one of 7 choices"
dob
"date of birth"
start_bp
"blood pressure at start - systolic/diastolic"
pre/post_wt_kg
"weight in kilograms at start/end"
start_mes
"Mayo endoscopic Score at start of treatment"
start_bss
"Bowel symptom score at start"
start_abd
"Abdominal symptom score at start"
start_sys
"Systemic symptom score at start"
start_coping
"Coping score at start"
start_emo
"Emotional symptom score at start"
start_dl
"Impact on Daily living score at start"
start_wbc
"White blood cell count in blood at start"
start_plt
"Platelet count in blood at start"
start_na
"Sodium level in serum at start"
start_k
"Potassium level in serum at start"
end_month
"Month of end visit"
end_day
"Day of end visit"
end_year
"Year of end visit"
end_mes
"Mayo endoscopic Score at end of treatment"
end_bss
"Bowel symptom score at end"
end_abd
"Abdominal symptom score at end"
end_sys
"Systemic symptom score at end"
end_coping
"Coping score at end"
end_emo
"Emotional symptom score at end"
end_dl
"Impact on Daily living score at end"
end_wbc
"White blood cell count in blood at end"
end_plt
"Platelet count in blood at end"
end_na
"Sodium level in serum at end"
end_k
"Potassium level in serum at end" # assign labels to the data set
df_clean <- df_raw |>
janitor::clean_names() |>
janitor::remove_empty(which = c("rows", "cols")) |>
mutate(
ethnic_clean = case_when(
ethnic %in% c("hispanic", "Hispanic", "hispamnic") ~ "hispanic",
ethnic %in% c("NOT hispanic", "not hispanic") ~ "not hispanic",
) |> fct_infreq(),
end_na_clean = na_if(end_na, -99),
end_emo_clean = na_if(end_emo, "not done") |> as.numeric(),
start_na_clean = parse_number(start_na),
treatment = fct_relevel(treatment, "upa", "uste", "oza")
) |>
separate_wider_delim(start_bp, delim ="/", names = c("bp_systolic", "bp_diastolic"), cols_remove = FALSE) |>
mutate(across(c(bp_systolic, bp_diastolic), as.numeric)) |>
# assign labels to all variables from the codebook
labelled::set_variable_labels(!!!vec_variables, .strict = FALSE) |>
# assign labels to new derived variables that did not exist in the code book
labelled::set_variable_labels(
ethnic_clean = "Ethnicity",
start_na_clean = "Sodium level in serum at start",
end_na_clean = "Sodium level in serum at end",
end_emo_clean = "Emotional symptom score at end",
bp_systolic = "Systolic blood pressure",
bp_diastolic = "Diastolic blood pressure"
)🤔 Why doesn’t pre_post_wt_kg have a label?
# view structure of data frame
df_clean |>
select(pat_id, start_na, start_na_clean, pre_post_wt_kg, start_emo) |>
str()tibble [30 × 5] (S3: tbl_df/tbl/data.frame)
$ pat_id : chr [1:30] "001" "002" "003" "004" ...
..- attr(*, "label")= chr "Patient Identifier"
$ start_na : chr [1:30] "137mmol/L" "142mmol/L" "140mmol/L" "139mmol/L" ...
..- attr(*, "label")= chr "Sodium level in serum at start"
$ start_na_clean: num [1:30] 137 142 140 139 144 145 142 138 140 137 ...
..- attr(*, "label")= chr "Sodium level in serum at start"
$ pre_post_wt_kg: chr [1:30] "84/82" "77/77" "74/75" "66/65" ...
$ start_emo : num [1:30] 79.3 58.6 32.2 75.6 74.3 ...
..- attr(*, "label")= chr "Emotional symptom score at start"Stage 2
Wrangling your Data
Where are We Now?
- We have done DEV (Data Exploration and Validation) - with more validation to be done. Data validation is a continuous process, right up to Data Lock.
- We discussed the importance of preventing data errors at the source - data collection and entry (preferably using a tool with data validation at entry - like REDCap)
- We did Stage 1 cleaning - clean variable names, remove empty columns/rows, fix variable types/classes (characters in numeric, recoding factors, date madness), address missingness, violations of tidy data principles (separate), added meaningful variable labels.
- While Stage 1 is required in nearly all projects, Stage 2 data cleaning is frequently needed, but not in all projects.
Stage 2 Data Cleaning
- We will cover these frequent but optional topics:
- Restructuring data as Long or Wide format
- Thickening or padding Longitudinal Data
- Joining multiple datasets
Restructuring data to Long or Wide format
Wide Format - One Row per Patient
# A tibble: 6 × 8
pat_id treatment start_mes end_mes start_bss end_bss start_emo end_emo
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 001 upa 3 0 75.9 9.85 79.3 39.8
2 002 uste 2 1 54.7 32.7 58.6 49.4
3 003 oza 1 1 28.5 28.5 32.2 35.9
4 004 upa 3 1 77.2 33.2 75.6 50.2
5 005 oza 3 2 78.8 56.8 74.3 65.0
6 006 uste 2 1 54.4 32.4 45.3 36.0Long Format - One Row per Measurement
# A tibble: 9 × 4
pat_id treatment measure score
<chr> <chr> <chr> <dbl>
1 001 upa start_mes 3
2 001 upa end_mes 0
3 001 upa start_bss 75.9
4 001 upa end_bss 9.85
5 001 upa start_emo 79.3
6 001 upa end_emo 39.8
7 002 uste start_mes 2
8 002 uste end_mes 1
9 002 uste start_bss 54.7 The Unit of Analysis - Wide by Patient
- We may want to do an analysis by patient, as each patient may (or may not) have the outcome. If we have multiple observations, or data points on each patient, this leads to wide data, with one row per patient.
# A tibble: 6 × 8
pat_id treatment start_mes end_mes start_bss end_bss start_emo end_emo
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 001 upa 3 0 75.9 9.85 79.3 39.8
2 002 uste 2 1 54.7 32.7 58.6 49.4
3 003 oza 1 1 28.5 28.5 32.2 35.9
4 004 upa 3 1 77.2 33.2 75.6 50.2
5 005 oza 3 2 78.8 56.8 74.3 65.0
6 006 uste 2 1 54.4 32.4 45.3 36.0The Unit of Analysis - Long by Visit
- We are often interested in the change in an outcome over time.
- To make this work, we need one row per measurement of the outcome. This leads to long data, with multiple visits and measurements for each patient.
# A tibble: 180 × 4
pat_id treatment measure score
<chr> <chr> <chr> <dbl>
1 001 upa start_mes 3
2 001 upa end_mes 0
3 001 upa start_bss 75.9
4 001 upa end_bss 9.85
5 001 upa start_emo 79.3
6 001 upa end_emo 39.8
7 002 uste start_mes 2
8 002 uste end_mes 1
9 002 uste start_bss 54.7
10 002 uste end_bss 32.7
# ℹ 170 more rowsDeciding on the Unit of Analysis
- This Unit of Analysis usually depends on the
Questionwe ask
Is the Unit of Analysis the Patient?
- Did the patient die?
- Did the patient have the outcome of colectomy?
- Did the patient reach disease remission?
Is the Unit of Analysis the Visit/Encounter?
- Often these are within-patient outcomes
- Did the C-reactive protein improve from Week 0 to Week 8?
- Did the sickle cell crises/year decrease after CRISPR gene Rx?
- Did the endoscopic score decrease on treatment vs placebo?
Deciding on the Unit of Analysis
- You will most often use long data, and this data structure allows you to look at multiple predictors and outcomes, like blood pressure, a PHQ-9 depression questionnaire, and a hemoglobin measurement.
- Depending on the analysis question, you may want to use wide data, and analyze by patient (usually with dichotomous outcomes).
Deciding on the Unit of Analysis
- For inpatient data, you may have multiple measurements in the same visit or day, so we need to decide a priori on how to handle multiple observations of the same type (e.g., vitals q6h) in the same day.
- use the
0600observation each day? - use the daily average for SBP and DBP?
- use the max values each day?
- use the
- Sometimes, you may want to do analysis on both long and wide data in the same project.
- It is often helpful to be able to pivot your data between long and wide structures.
Reshaping your data with tidyr
- We often enter data by patient
- Spreadsheets encourage us to enter longitudinal data as long rows (per patient)
- We end up with
wide, rather thantalldata
# A tibble: 6 × 8
pat_id treatment start_mes end_mes start_bss end_bss start_emo end_emo
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 001 upa 3 0 75.9 9.85 79.3 39.8
2 002 uste 2 1 54.7 32.7 58.6 49.4
3 003 oza 1 1 28.5 28.5 32.2 35.9
4 004 upa 3 1 77.2 33.2 75.6 50.2
5 005 oza 3 2 78.8 56.8 74.3 65.0
6 006 uste 2 1 54.4 32.4 45.3 36.0# A tibble: 180 × 4
pat_id treatment measure score
<chr> <chr> <chr> <dbl>
1 001 upa start_mes 3
2 001 upa end_mes 0
3 001 upa start_bss 75.9
4 001 upa end_bss 9.85
5 001 upa start_emo 79.3
6 001 upa end_emo 39.8
7 002 uste start_mes 2
8 002 uste end_mes 1
9 002 uste start_bss 54.7
10 002 uste end_bss 32.7
# ℹ 170 more rowsReshaping your data with tidyr
- R (and most R functions) are vectorized to handle tall data
- One small observation per row
- Most analyses in R are easier with tall data
# A tibble: 180 × 4
pat_id treatment measure score
<chr> <chr> <chr> <dbl>
1 001 upa start_mes 3
2 001 upa end_mes 0
3 001 upa start_bss 75.9
4 001 upa end_bss 9.85
5 001 upa start_emo 79.3
6 001 upa end_emo 39.8
7 002 uste start_mes 2
8 002 uste end_mes 1
9 002 uste start_bss 54.7
10 002 uste end_bss 32.7
# ℹ 170 more rowsPivoting Longer (common)
- We need to ‘pivot’ data from wide to tall on the regular
- This “lengthens” data, increasing the number of rows, and decreasing the number of columns
- We will be looking at Visit Dates (Start vs End) and Measures
Pivoting Longer
- Arguments:
data,cols,names_to,values_to, and many optional arguments - Details from the tidyverse help page are here
data= your dataframe/tibble - you can pipe this incols= columns to pivot, as a vector of names, or by number, or selected with tidyselect functionsnames_to= A character vector specifying the new column or columns to create from the information stored in the column names of data specified by cols.values_to= A string specifying the name of the column to create from the data stored in cell values.
Pivoting Longer (Example)
Let’s start with the wide version (selected columns from messy_uc)
# A tibble: 30 × 8
pat_id treatment start_mes end_mes start_bss end_bss start_emo end_emo
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 001 upa 3 0 75.9 9.85 79.3 39.8
2 002 uste 2 1 54.7 32.7 58.6 49.4
3 003 oza 1 1 28.5 28.5 32.2 35.9
4 004 upa 3 1 77.2 33.2 75.6 50.2
5 005 oza 3 2 78.8 56.8 74.3 65.0
6 006 uste 2 1 54.4 32.4 45.3 36.0
7 007 uste 3 2 79.7 57.7 66.6 57.8
8 008 oza 3 2 75.0 53.0 74.5 62.0
9 009 upa 2 0 53.1 9.06 50.0 27.8
10 010 oza 3 1 78.9 34.9 72.5 45.9
# ℹ 20 more rows- Note that there are 30 rows, one per patient, with 6 measured quantities for each patient.
Pivoting Longer (Example)
This is the tall version we want to end up with.
# A tibble: 180 × 4
pat_id treatment measure score
<chr> <chr> <chr> <dbl>
1 001 upa start_mes 3
2 001 upa end_mes 0
3 001 upa start_bss 75.9
4 001 upa end_bss 9.85
5 001 upa start_emo 79.3
6 001 upa end_emo 39.8
7 002 uste start_mes 2
8 002 uste end_mes 1
9 002 uste start_bss 54.7
10 002 uste end_bss 32.7
# ℹ 170 more rows- Note that there now 180 rows (30*6), with one row per observation measure.
Doing the pivot_longer()
What values do we want for these key arguments in order to pivot_longer?
cols(which columns to pivot?)names_to(variable to store the names)values_to(variable to store the values)
# A tibble: 30 × 8
pat_id treatment start_mes end_mes start_bss end_bss start_emo end_emo
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 001 upa 3 0 75.9 9.85 79.3 39.8
2 002 uste 2 1 54.7 32.7 58.6 49.4
3 003 oza 1 1 28.5 28.5 32.2 35.9
4 004 upa 3 1 77.2 33.2 75.6 50.2
5 005 oza 3 2 78.8 56.8 74.3 65.0
6 006 uste 2 1 54.4 32.4 45.3 36.0
7 007 uste 3 2 79.7 57.7 66.6 57.8
8 008 oza 3 2 75.0 53.0 74.5 62.0
9 009 upa 2 0 53.1 9.06 50.0 27.8
10 010 oza 3 1 78.9 34.9 72.5 45.9
# ℹ 20 more rowsPivoting Longer In Action
# A tibble: 30 × 8
pat_id treatment start_mes end_mes start_bss end_bss start_emo end_emo
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 001 upa 3 0 75.9 9.85 79.3 39.8
2 002 uste 2 1 54.7 32.7 58.6 49.4
3 003 oza 1 1 28.5 28.5 32.2 35.9
4 004 upa 3 1 77.2 33.2 75.6 50.2
5 005 oza 3 2 78.8 56.8 74.3 65.0
6 006 uste 2 1 54.4 32.4 45.3 36.0
7 007 uste 3 2 79.7 57.7 66.6 57.8
8 008 oza 3 2 75.0 53.0 74.5 62.0
9 009 upa 2 0 53.1 9.06 50.0 27.8
10 010 oza 3 1 78.9 34.9 72.5 45.9
# ℹ 20 more rows1wide |>
pivot_longer(
cols = "start_mes":"end_emo",
2 names_to = "measure",
3 values_to = "score"
4 )- 1
- start with wide data, and pivot_longer
- 2
- Which columns to pivot (names in quotes), could also use 3:8
- 3
- which column (quotes) the pivoted variable names should go into
- 4
- which column (quotes) the pivoted values should go into
# A tibble: 180 × 4
pat_id treatment measure score
<chr> <chr> <chr> <dbl>
1 001 upa start_mes 3
2 001 upa end_mes 0
3 001 upa start_bss 75.9
4 001 upa end_bss 9.85
5 001 upa start_emo 79.3
6 001 upa end_emo 39.8
7 002 uste start_mes 2
8 002 uste end_mes 1
9 002 uste start_bss 54.7
10 002 uste end_bss 32.7
# ℹ 170 more rows- Does this make sense so far?
- Zoom reaction: “thumbs up” emoji 👍 if yes
- “raised hand” emoji ✋ if puzzled/questions
One Minor Issue - Separation of measure
- the “measure” column combines a timepoint and the measure
- Needs to be separated.
- You already know how to use separate()
- Arguments
colsepinto
# A tibble: 180 × 5
pat_id treatment timept measure score
<chr> <chr> <chr> <chr> <dbl>
1 001 upa start mes 3
2 001 upa end mes 0
3 001 upa start bss 75.9
4 001 upa end bss 9.85
5 001 upa start emo 79.3
6 001 upa end emo 39.8
7 002 uste start mes 2
8 002 uste end mes 1
9 002 uste start bss 54.7
10 002 uste end bss 32.7
# ℹ 170 more rows- You can do this within pivot_longer with just one more argument
(if you read all of the pivot_longer documentation)
# A tibble: 180 × 5
pat_id treatment timept measure score
<chr> <chr> <chr> <chr> <dbl>
1 001 upa start mes 3
2 001 upa end mes 0
3 001 upa start bss 75.9
4 001 upa end bss 9.85
5 001 upa start emo 79.3
6 001 upa end emo 39.8
7 002 uste start mes 2
8 002 uste end mes 1
9 002 uste start bss 54.7
10 002 uste end bss 32.7
# ℹ 170 more rowsPivoting Longer
- Your Turn with
endo_data - Measurements of Trans-Epithelial Electrical Resistance (TEER, the inverse of leakiness) in biopsies of 3 segments of intestine.
- This
couldbe affected by portal hypertension in patients with liver cirrhosis - Let’s find out!
Doing this At Home? Data load
- Here is the code to load the data if you are doing this on a local computer. Use the clipboard icon at the top right to copy the code.
endo_data <- tibble::tribble(
~pat_id, ~portal_htn, ~duod_teer, ~ileal_teer, ~colon_teer,
001, 1, 4.33, 14.57, 16.23,
002, 0, 11.67, 15.99, 18.97,
003, 1, 4.12, 13.77, 15.22,
004, 1, 4.62, 16.37, 18.12,
005, 0, 12.43, 15.84, 19.04,
006, 0, 13.05, 16.23, 18.81,
007, 0, 11.88, 15.72, 18.31,
008, 1, 4.87, 16.59, 18.77,
009, 1, 4.23, 15.04, 16.87,
010, 0, 12.77, 16.73, 19.12
)
endo_data# A tibble: 10 × 5
pat_id portal_htn duod_teer ileal_teer colon_teer
<dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 4.33 14.6 16.2
2 2 0 11.7 16.0 19.0
3 3 1 4.12 13.8 15.2
4 4 1 4.62 16.4 18.1
5 5 0 12.4 15.8 19.0
6 6 0 13.0 16.2 18.8
7 7 0 11.9 15.7 18.3
8 8 1 4.87 16.6 18.8
9 9 1 4.23 15.0 16.9
10 10 0 12.8 16.7 19.1Exercise PH1
Complete Data Cleaning Fundamentals Exercise PH1.
Do This in Posit Cloud
If you have the exercise done correctly, click on the Reactions tab in Zoom, and click to put the “thumbs up” emoji 👍 on your screen.
If you are having trouble, click on the Reactions tab in Zoom, and click to put the “raised hand” emoji ✋ on your screen.
–> Take me to the exercise Solution <–
03:00
Pivoting Longer with endo_data
# A tibble: 10 × 5
pat_id portal_htn duod_teer ileal_teer colon_teer
<dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 4.33 14.6 16.2
2 2 0 11.7 16.0 19.0
3 3 1 4.12 13.8 15.2
4 4 1 4.62 16.4 18.1
5 5 0 12.4 15.8 19.0
6 6 0 13.0 16.2 18.8
7 7 0 11.9 15.7 18.3
8 8 1 4.87 16.6 18.8
9 9 1 4.23 15.0 16.9
10 10 0 12.8 16.7 19.1- What values do you want to use for these arguments to
pivot_longer:colsnames_pattern= “(.+)_teer”names_tovalues_to
- Note that we are giving you the correct value for
names_pattern, which will ask for what we want - to keep the characters of the name (of whatever length) before “_teer”
- Fill in the blanks to pivot this dataset to tall format, with columns for the intestinal location and the teer value.
- Note that we are giving you the correct answer for the
names_patternargument.
- Fill in the blanks to pivot this dataset to tall format, with columns for the intestinal location and the teer value.
- Run the code, and look at the resulting table. Use the clipboard icon at the top right to copy the code.
# A tibble: 30 × 4
pat_id portal_htn location teer
<dbl> <dbl> <chr> <dbl>
1 1 1 duod 4.33
2 1 1 ileal 14.6
3 1 1 colon 16.2
4 2 0 duod 11.7
5 2 0 ileal 16.0
6 2 0 colon 19.0
7 3 1 duod 4.12
8 3 1 ileal 13.8
9 3 1 colon 15.2
10 4 1 duod 4.62
# ℹ 20 more rows- Do you think that portal hypertension has an effect on TEER and (its inverse) epithelial leakiness?
Pivoting Wider
- Wide data is less common, but sometimes needed for per-patient analysis
- Here we will convert the tall version of our selected messy_uc data back to wide.
- This is what the tall data look like
# A tibble: 180 × 4
pat_id treatment measure score
<chr> <chr> <chr> <dbl>
1 001 upa start_mes 3
2 001 upa end_mes 0
3 001 upa start_bss 75.9
4 001 upa end_bss 9.85
5 001 upa start_emo 79.3
6 001 upa end_emo 39.8
7 002 uste start_mes 2
8 002 uste end_mes 1
9 002 uste start_bss 54.7
10 002 uste end_bss 32.7
# ℹ 170 more rows# A tibble: 30 × 8
pat_id treatment start_mes end_mes start_bss end_bss start_emo end_emo
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 001 upa 3 0 75.9 9.85 79.3 39.8
2 002 uste 2 1 54.7 32.7 58.6 49.4
3 003 oza 1 1 28.5 28.5 32.2 35.9
4 004 upa 3 1 77.2 33.2 75.6 50.2
5 005 oza 3 2 78.8 56.8 74.3 65.0
6 006 uste 2 1 54.4 32.4 45.3 36.0
7 007 uste 3 2 79.7 57.7 66.6 57.8
8 008 oza 3 2 75.0 53.0 74.5 62.0
9 009 upa 2 0 53.1 9.06 50.0 27.8
10 010 oza 3 1 78.9 34.9 72.5 45.9
# ℹ 20 more rowsLongitudinal Data
- Another set of data issues you may face is in data collected over time, when one of two things happen:
- You want to analyze data by day or month or year, and data in your Electronic Medical Record is collected and time-stamped by the
second. - You realize that some observations (on weekends) are missing, and you need to fill these dates in as missing, but you really don’t want to do this by hand.
- You want to analyze data by day or month or year, and data in your Electronic Medical Record is collected and time-stamped by the
- The {padr} package can help with these issues.

Thicken Date-Times with padr
# A tibble: 6 × 2
title time_stamp
<chr> <dttm>
1 EMS: BACK PAINS/INJURY 2015-12-10 17:40:00
2 EMS: DIABETIC EMERGENCY 2015-12-10 17:40:00
3 Fire: GAS-ODOR/LEAK 2015-12-10 17:40:00
4 EMS: CARDIAC EMERGENCY 2015-12-10 17:40:01
5 EMS: DIZZINESS 2015-12-10 17:40:01
6 EMS: HEAD INJURY 2015-12-10 17:40:01- The emergency data set in the {padr} package contains > 120K emergency calls from Montgomery County, PA over a period of ~ 11 months.
- Each call has a title and a timestamp
- We want to know in which months the emergency department should order an extra case of Narcan
(H: summer months?).
Thickening Time to a Usable Level
- The
thickenfunction adds a column to a data frame that is of a higher interval than the original variable. - The intervals for {padr} are year, quarter, month, week, day, hour, min, and sec.
- The variable
time_stamphas the interval of seconds - We can thicken the data to the time interval we need.
- Then we can count events by a usable unit of time
# A tibble: 6 × 6
lat lng zip title time_stamp twp
<dbl> <dbl> <int> <chr> <dttm> <chr>
1 40.3 -75.6 19525 EMS: BACK PAINS/INJURY 2015-12-10 17:40:00 NEW HANOVER
2 40.3 -75.3 19446 EMS: DIABETIC EMERGENCY 2015-12-10 17:40:00 HATFIELD TOWNSH…
3 40.1 -75.4 19401 Fire: GAS-ODOR/LEAK 2015-12-10 17:40:00 NORRISTOWN
4 40.1 -75.3 19401 EMS: CARDIAC EMERGENCY 2015-12-10 17:40:01 NORRISTOWN
5 40.3 -75.6 NA EMS: DIZZINESS 2015-12-10 17:40:01 LOWER POTTSGROVE
6 40.3 -75.3 19446 EMS: HEAD INJURY 2015-12-10 17:40:01 LANSDALE - We will thicken to month
# A tibble: 6 × 4
title time_stamp twp time_stamp_month
<chr> <dttm> <chr> <date>
1 EMS: BACK PAINS/INJURY 2015-12-10 17:40:00 NEW HANOVER 2015-12-01
2 EMS: DIABETIC EMERGENCY 2015-12-10 17:40:00 HATFIELD TOWNSHIP 2015-12-01
3 Fire: GAS-ODOR/LEAK 2015-12-10 17:40:00 NORRISTOWN 2015-12-01
4 EMS: CARDIAC EMERGENCY 2015-12-10 17:40:01 NORRISTOWN 2015-12-01
5 EMS: DIZZINESS 2015-12-10 17:40:01 LOWER POTTSGROVE 2015-12-01
6 EMS: HEAD INJURY 2015-12-10 17:40:01 LANSDALE 2015-12-01 - We will thicken to week
# A tibble: 6 × 4
title time_stamp twp time_stamp_week
<chr> <dttm> <chr> <date>
1 EMS: BACK PAINS/INJURY 2015-12-10 17:40:00 NEW HANOVER 2015-12-06
2 EMS: DIABETIC EMERGENCY 2015-12-10 17:40:00 HATFIELD TOWNSHIP 2015-12-06
3 Fire: GAS-ODOR/LEAK 2015-12-10 17:40:00 NORRISTOWN 2015-12-06
4 EMS: CARDIAC EMERGENCY 2015-12-10 17:40:01 NORRISTOWN 2015-12-06
5 EMS: DIZZINESS 2015-12-10 17:40:01 LOWER POTTSGROVE 2015-12-06
6 EMS: HEAD INJURY 2015-12-10 17:40:01 LANSDALE 2015-12-06 Thickening Time for a Monthly Plot
- The thicken function adds a column to a data frame that is of a higher interval than the original variable.
- The variable
time_stamphas the interval of seconds - We want to thicken this (time stamped) data to month, then select and count overdoses
- This will set up the monthly overdose plot we want.
- We will thicken to month
- Then count overdoses by month
- This lets us count events like overdoses by month with time_stamp_month.
# A tibble: 11 × 2
time_stamp_month overdoses
<date> <int>
1 2015-12-01 91
2 2016-01-01 135
3 2016-02-01 141
4 2016-03-01 161
5 2016-04-01 124
6 2016-05-01 149
7 2016-06-01 140
8 2016-07-01 147
9 2016-08-01 137
10 2016-09-01 171
11 2016-10-01 95
- When would you order extra cases of Narcan?
Padding unobserved dates (weekends?)
- The pad() function allows you to fill in missing intervals.
- As an example, my hospital only runs fecal calprotectin tests on weekdays.
- This can lead to weird discontinuities in data over a weekend (Dec 3-4).
- No observations on weekend days/holidays.
# A tibble: 14 × 3
pat_id date fcp
<chr> <date> <dbl>
1 001 2022-12-01 1574
2 001 2022-12-02 1323
3 001 2022-12-05 673
4 001 2022-12-06 314
5 001 2022-12-07 168
6 002 2022-11-30 1393
7 002 2022-12-01 1014
8 002 2022-12-02 812
9 002 2022-12-05 247
10 002 2022-12-06 118
11 003 2022-12-02 987
12 003 2022-12-05 438
13 003 2022-12-06 312
14 003 2022-12-05 194Padding Unobserved Times
- We can fill in (pad) the unobserved weekend days with the pad() function.
# A tibble: 21 × 3
pat_id date fcp
<chr> <date> <dbl>
1 001 2022-12-01 1574
2 001 2022-12-02 1323
3 001 2022-12-03 NA
4 001 2022-12-04 NA
5 001 2022-12-05 673
6 001 2022-12-06 314
7 001 2022-12-07 168
8 002 2022-11-30 1393
9 002 2022-12-01 1014
10 002 2022-12-02 812
11 002 2022-12-03 NA
12 002 2022-12-04 NA
# ℹ 9 more rows- New observations are created on the missing dates
- NAs are filled in for the missing FCPs, with one for each day and group (pat_id)
- pad() fills in the missing pat_ids
Joining Data
- Another data issue you will often face is that you have two interesting data sets, and that they would be more interesting if you could link the data in one to the data in the other.

Chocolate and Peanut Butter
(Datasets)
Joins of data from different sources
- We often collect data from different sources that we later want to join together for analysis
- Data from your local Electronic Medical Record
- Data from the CDC
- Data from the US Census
- External data can illuminate our understanding of our local patient data

Local Demographics with CDC SVI data
- We have 2 datasets, one local Demographics and Census Tract, and one from the CDC that has values for Social Vulnerability Index by Census Tract
- We want to know if the SVI for the neighborhood of each patient influences health outcomes
- We need to
left_jointhese datasets together by matching on the Census Tract
- What is the common uniqid/key?
- Local EMR data (
demo)
# A tibble: 9 × 4
pat_id name htn census_tract
<chr> <chr> <dbl> <dbl>
1 001 Arthur Blankenship 0 26161404400
2 002 Britney Jonas 0 26161405100
3 003 Sally Davis 1 26161402100
4 004 Al Jones 0 26161403200
5 005 Gary Hamill 1 26161405200
6 006 Ken Bartoletti 0 26161404500
7 007 Ike Gerhold 0 26161405600
8 008 Tatiana Grant 0 26161404300
9 009 Antione Delacroix 1 26161405500cdc
# A tibble: 9 × 2
census_tract svi
<dbl> <dbl>
1 26161404400 0.12
2 26161405100 0.67
3 26161402100 0.43
4 26161403200 0.07
5 26161405200 0.71
6 26161404500 0.23
7 26161405600 0.27
8 26161404300 0.21
9 26161405500 0.62- Replace the generic arguments to the left_join function to join the demographic data (demo) on the left to add Social Vulnerability index (svi) from the cdc dataset on the RHS. Left join by census tract.
- Now that the join is complete, you can ask your question:
- Is there an association between social vulnerability and hypertension?
# A tibble: 9 × 5
pat_id name htn census_tract svi
<chr> <chr> <dbl> <dbl> <dbl>
1 001 Arthur Blankenship 0 26161404400 0.12
2 002 Britney Jonas 0 26161405100 0.67
3 003 Sally Davis 1 26161402100 0.43
4 004 Al Jones 0 26161403200 0.07
5 005 Gary Hamill 1 26161405200 0.71
6 006 Ken Bartoletti 0 26161404500 0.23
7 007 Ike Gerhold 0 26161405600 0.27
8 008 Tatiana Grant 0 26161404300 0.21
9 009 Antione Delacroix 1 26161405500 0.62Patient Demographics with Lab results (Your Turn to Join)
- We have some basic Patient Demographics in one table (demo)
# A tibble: 9 × 3
pat_id name age
<chr> <chr> <dbl>
1 001 Arthur Blankenship 67
2 002 Britney Jonas 23
3 003 Sally Davis 63
4 004 Al Jones 44
5 005 Gary Hamill 38
6 006 Ken Bartoletti 33
7 007 Ike Gerhold 52
8 008 Tatiana Grant 42
9 009 Antione Delacroix 27and potassium levels and creatinine levels in 2 other tables (pot and cr)
# A tibble: 5 × 2
pat_id k
<chr> <dbl>
1 001 3.2
2 002 3.7
3 003 4.2
4 004 4.4
5 005 4.1# A tibble: 5 × 2
pat_id cr
<chr> <dbl>
1 001 0.2
2 002 0.5
3 003 0.9
4 004 1.5
5 005 0.7Need to Load the Data?
If you are trying this on your local computer, copy the code below with the clipboard icon to get the data into your computer.
demo <- tibble::tribble(
~pat_id, ~name, ~age,
'001', "Arthur Blankenship", 67,
'002', "Britney Jonas", 23,
'003', "Sally Davis", 63,
'004', "Al Jones", 44,
'005', "Gary Hamill", 38,
'006', "Ken Bartoletti", 33,
'007', "Ike Gerhold", 52,
'008', "Tatiana Grant", 42,
'009', "Antoine Delacroix", 27,
)
pot <- tibble::tribble(
~pat_id, ~k,
'001', 3.2,
'002', 3.7,
'003', 4.2,
'004', 4.4,
'005', 4.1,
'006', 4.0,
'007', 3.6,
'008', 4.2,
'009', 4.9,
)
cr <- tibble::tribble(
~pat_id, ~cr,
'001', 0.2,
'002', 0.5,
'003', 0.9,
'004', 1.5,
'005', 0.7,
'006', 0.9,
'007', 0.7,
'008', 1.0,
'009', 1.7,
)Your Turn to Join
- We want to join the correct labs (9 rows each from the pot and cr datasets) to the correct patients.
- The unique identifier (called the uniqid or key or recordID) is pat_id.
- It only occurs once for each patient/row
- It appears in each table we want to join
- The pat_id is of the character type in each (a common downfall if one is character, one is numeric, but they look the same - but don’t match)
- We want to start with demographics, then add datasets that match to the right.
- We will use demo as our base dataset on the left hand side (LHS), and first join the potassium (pot) results (RHS)
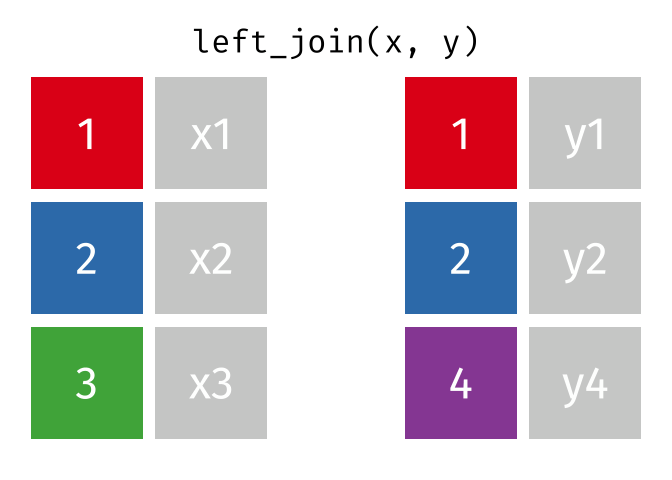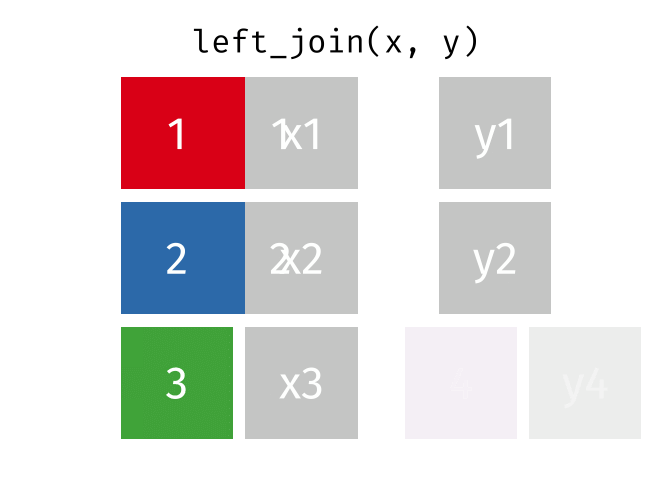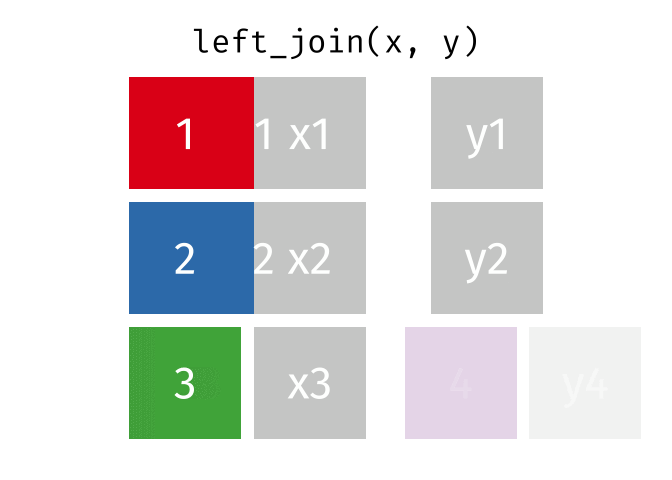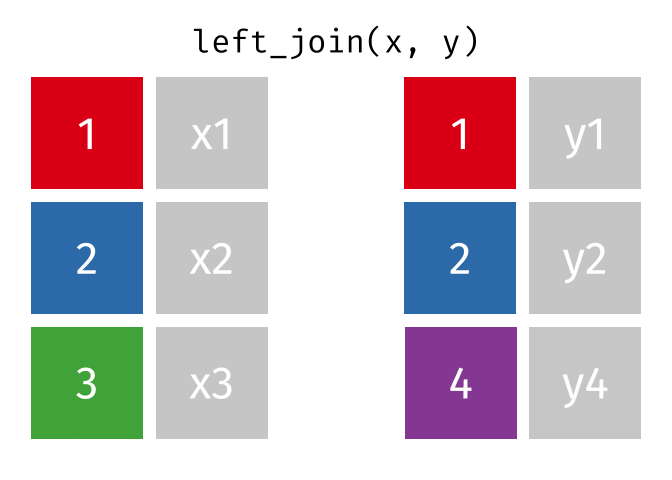
What the Left Join Looks Like
This is a mutating join - new variables from y are created/added to the LHS (x).
Exercise PH2
Complete the Data Cleaning Fundamentals Exercise PH2.
Do This in Posit Cloud
If you have the exercise done correctly, click on the Reactions tab in Zoom, and click to put the “thumbs up” emoji 👍 on your screen.
If you are having trouble, click on the Reactions tab in Zoom, and click to put the “raised hand” emoji ✋ on your screen.
–> Take me to the exercise Solution <–
03:00
Your Turn to Join
- Joining demo to pot with a left_join
- left_join(
data_x,data_y, by ="uniqid")
- replace the generic arguments below with the correct ones to join demo to pot and produce new_data.
Note the syntax for identifying the uniqid on which to do the merge: by = "varname"
# A tibble: 9 × 4
pat_id name age k
<chr> <chr> <dbl> <dbl>
1 001 Arthur Blankenship 67 3.2
2 002 Britney Jonas 23 3.7
3 003 Sally Davis 63 4.2
4 004 Al Jones 44 4.4
5 005 Gary Hamill 38 4.1
6 006 Ken Bartoletti 33 4
7 007 Ike Gerhold 52 3.6
8 008 Tatiana Grant 42 4.2
9 009 Antoine Delacroix 27 4.9Now add Creatinine (cr) to new_data
- Joining new_data and cr with a
left_join - left_join(data_x, data_y, by = “uniqid”)
- Replace the generic arguments with the correct ones to join
new_data and cr and produce new_data2.
# A tibble: 9 × 5
pat_id name age k cr
<chr> <chr> <dbl> <dbl> <dbl>
1 001 Arthur Blankenship 67 3.2 0.2
2 002 Britney Jonas 23 3.7 0.5
3 003 Sally Davis 63 4.2 0.9
4 004 Al Jones 44 4.4 1.5
5 005 Gary Hamill 38 4.1 0.7
6 006 Ken Bartoletti 33 4 0.9
7 007 Ike Gerhold 52 3.6 0.7
8 008 Tatiana Grant 42 4.2 1
9 009 Antoine Delacroix 27 4.9 1.7- Al has HTN and DM2
- Antoine has early stage FSGS
Workhorse Joins
- left_join is your workhorse. Start with patient identifiers/uniqid and add data to the right side.
- Sometimes you will need to wrangle/process incoming data, then pipe it in to right_join it to the patient demographics

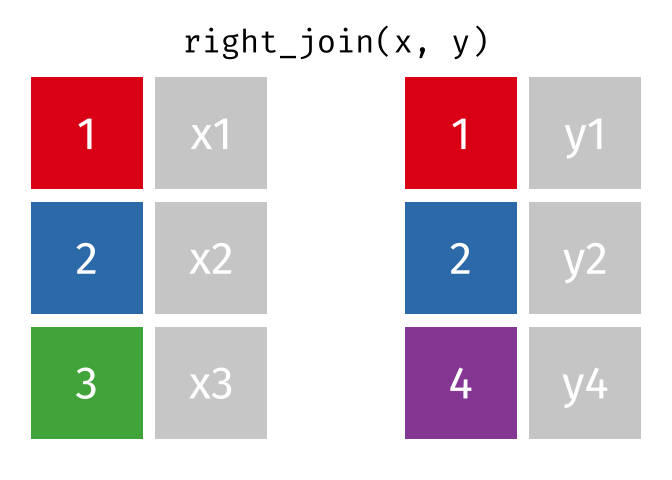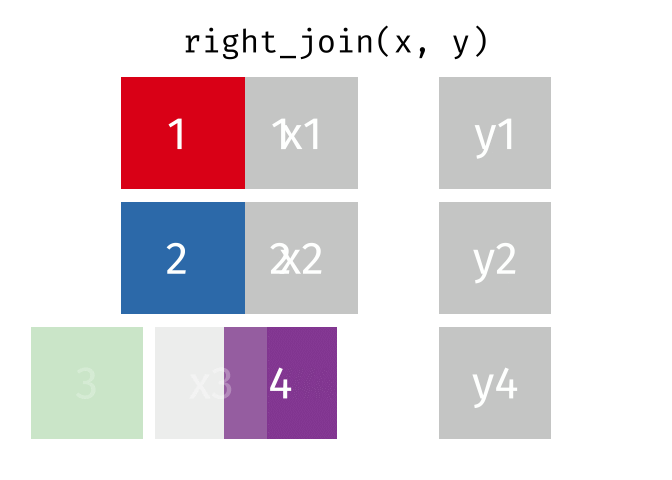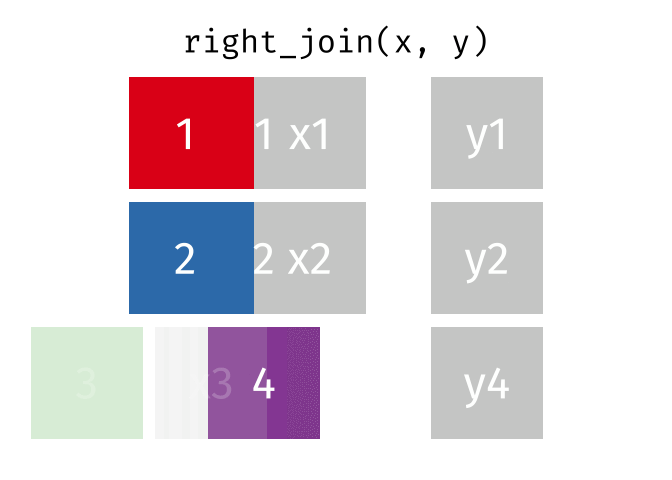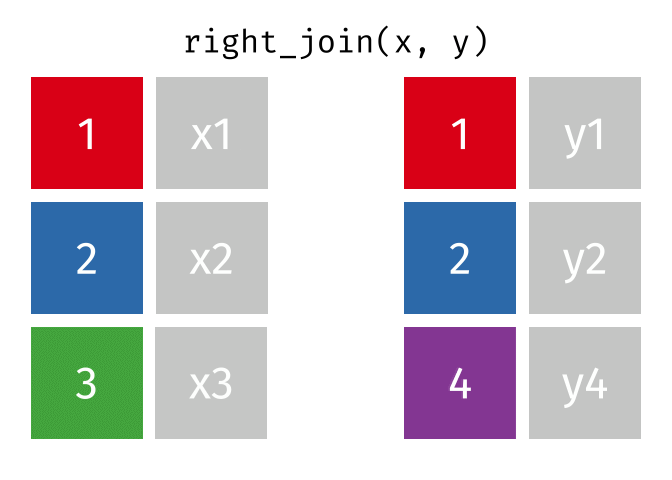
Fancy Joins
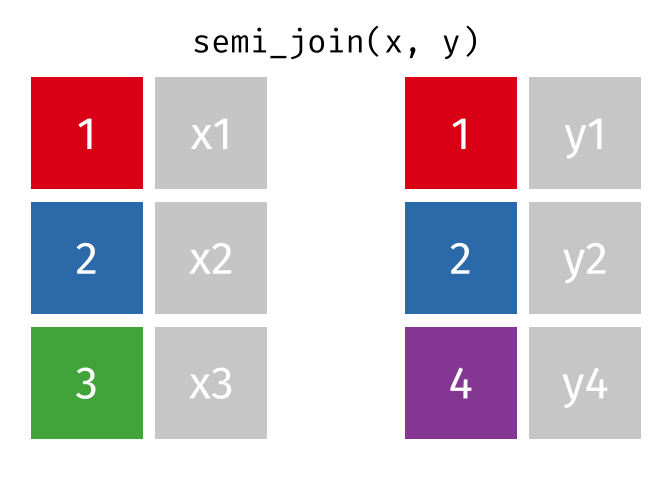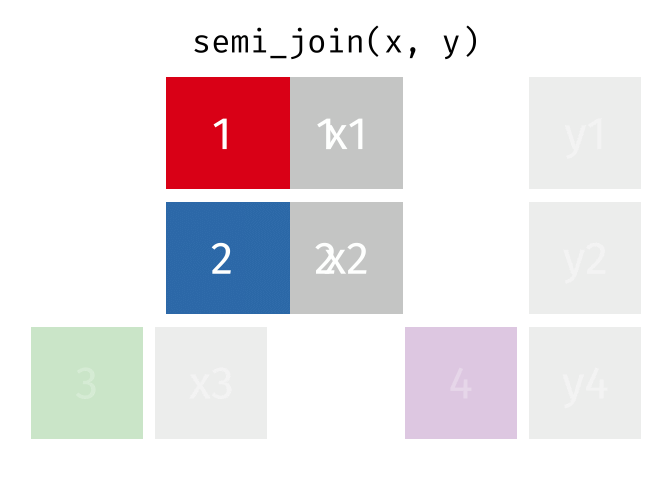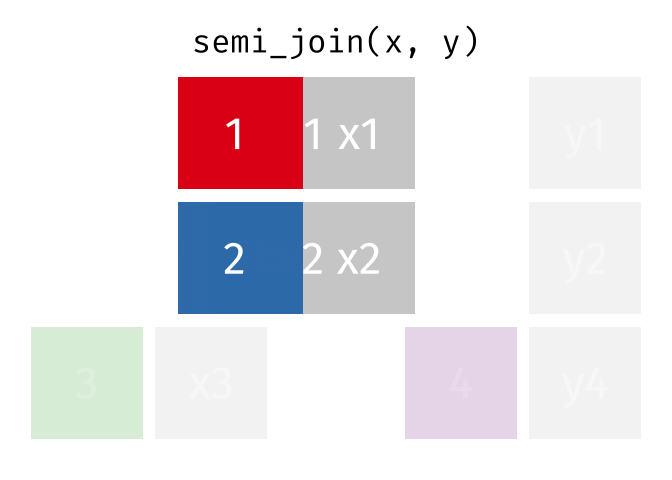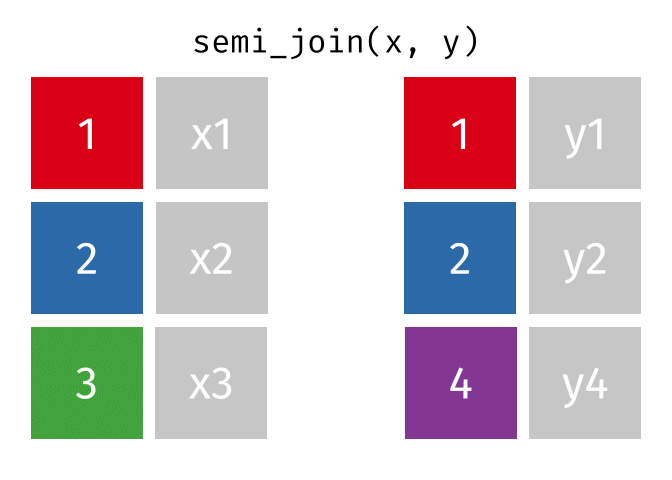
- There are multiple kinds of fancy joins (semi_join, anti_join, inner_join, full_join, union, intersect, setdiff) which come in handy once in a while.
- The many kinds of joins are all well explained here
- Subset your data (x), keeping only the ones (y=hospitalized) who were hospitalized.
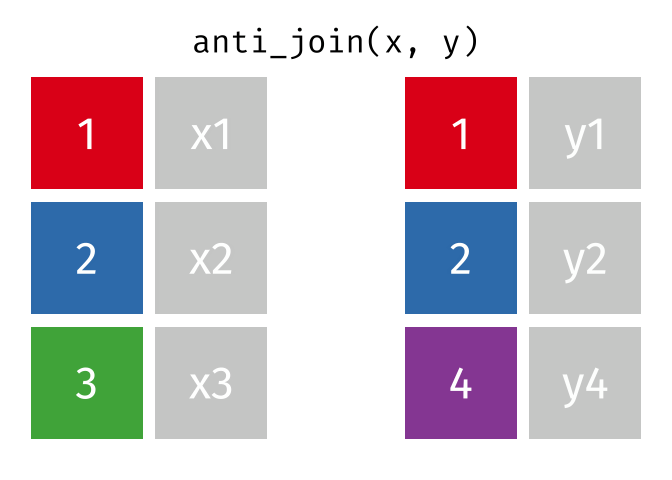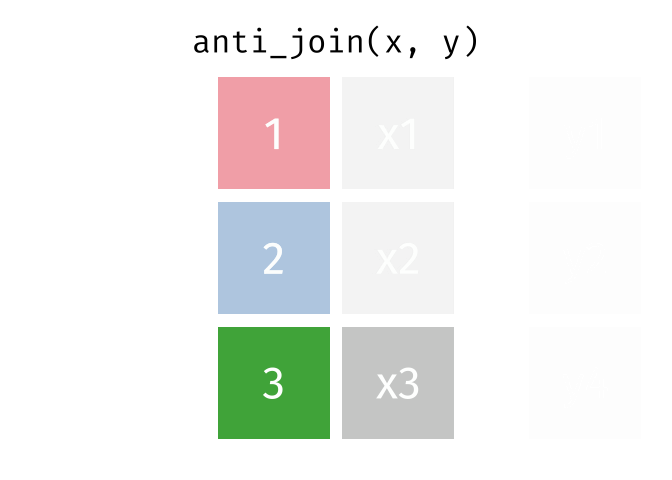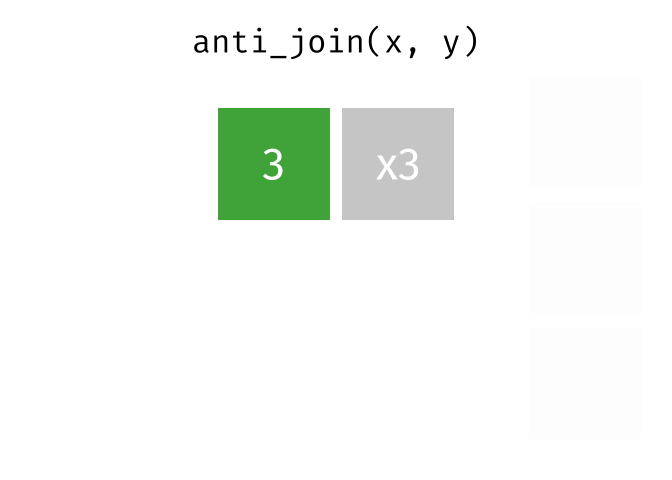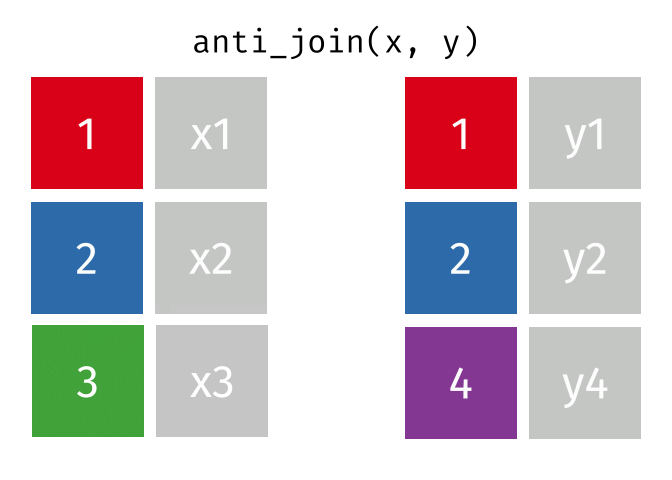
- Subset your data (x), keeping only the ones (y = dead) who are NOT dead.
The Newest Fancy Joins
- You can use
join_byfor more joining options, including- inequality - only match if > value
- closest date
closestdate as long it is after the comparison date, but within 30 days- only match if interval/date
overlaps - match value if it is
withina range
- Learn more at Join Specifications
Data Horror
Data So Messy, it Constitutes a Data Crime

What Should You Do?
Two Options
- You can find the person who collected/entered the data
- If you don’t correct this behavior now, this will torture many future data analysts
- Educate in this teachable moment about tidy data
- Send them to watch Tidy Spreadsheets on YouTube here to prevent this kind of Data Crime in the future.
- Improve the world, one (now tidy) data collector at a time.
- There is no way to find the person who collected/entered the data
- Pull out some advanced data cleaning packages made for this particular kind of mess
- {unpivotr}
- {tidyxl}
- {unheadr}
- Watch the unpivotr/tidyxl 14 min video here
- Learn from a free e-book, Spreadsheet Munging Strategies
Step by Step Cleaning - Stage 1
- Import your data cleanly - skip rows, get headers
- Explore your data
- Remove empty rows & columns
- Fix variable classes/types, check dates
- Clean variable names
- Create a codebook
- Validate for missingness, outliers
- Fix bad category values
- Identify and consistently label missing values
- Separate mixed (SBP/DBP) values, assign variable labels
Step by Step Cleaning - Stage 2 Wrangling
- Let your Question define your Unit of Analysis
- Pivot_longer or pivot_wider if needed
- Thicken time, pad missing times if needed
- Join to add new data if needed
- Know what packages to use for data horrors
- unpivotr
- tidyxl
- unheadr
Thank you!
Thank you for joining the workshop!
All materials will live on here: https://shannonpileggi.github.io/rmedicine-data-cleaning-2023/


